全文HTML
--> --> --> 炼焦企业为工业废水排放大户。每年焦化企业排放废水量达到4.3×108 m3,焦化废水COD排放量约为3.29×104 m3,总氮排放量约为8.2×104 m3,极大威胁了企业周边的水环境[1]。焦化废水为典型的高含氮难降解工业废水,主流的生化处理工艺为AO(缺氧-好氧)、AAO(厌氧-缺氧-好氧)和多级AO等[2]。通常,主流工艺利用微生物氨化、硝化和反硝化作用进行生物脱氮。焦化废水生化工艺结束后,在后续的焦化废水深度处理工艺中,压力驱动的纳滤膜技术已成功应用于生产实践,实现了焦化废水回用的目的[3]。唐山化工焦化废水系统年处理量6×105 m3,采用纳滤(nanofiltration,NF)工艺后,废水回收率大于75%[4]。在纳滤工艺处理焦化废水的过程中,纳滤膜分离出来的氯离子、硫酸根离子、硝酸根离子和含氮杂环有机物等均留在纳滤浓水中。有研究[5]表明,焦化纳滤浓水为高含盐、高含氮和高COD的“三高”废水,其中,总氮(total nitrogen,TN)主要由硝态氮(本研究以焦化纳滤工艺中产生的高电导率浓水为研究对象,通过投加外加碳源(甲醇),考察序批式反应器(sequencing batch reactor,SBR)对焦化浓水中总氮和硝态氮的去除效果;同时,采用高通量测序技术和qPCR定量技术深入分析纳滤浓水生物脱氮过程中微生物菌群结构和反硝化功能基因变化,以期为优化生物反应器的运行提供参考。
1.1. 实验原料、装置与方法
实验用的焦化纳滤浓水取自某钢厂焦化纳滤系统。焦化纳滤浓水水质变化特性如下:pH为7.2~7.9,电导率为9 450~11 250 μS·cm?1,COD为318~496 mg·L?1,BOD5为16~38 mg·L?1,TN为49~66 mg·L?1,实验采用的SBR装置是一个内径为14.0 cm、高为28.5 cm的敞口反应器,总容积为6.0 L,有效容积为5.0 L。SBR均由机械搅拌系统、曝气系统、进出水蠕动泵系统以及时间控制系统组成。SBR实验装置运行参数如下:进水时间为15 min,缺氧搅拌时间为6 h,好氧曝气时间为10 h,沉淀时间为30 min,出水时间为15 min,闲置时间为7 h,驯化周期为24 h,排出比为1∶2。
SBR启动的接种污泥取自焦化废水生化系统缺氧段,初始混合液悬浮固体浓度(mixed liquor suspended solids,MLSS)为4 100 mg·L?1。在驯化期间,纳滤浓水和焦化废水比例分别为60%∶40%、80%∶20%和100%∶0%;每个混合液比例条件下依次运行10 d,驯化期共30 d,不排泥。驯化完成后,稳定运行了60 d,反应器按时排泥,泥龄(sludge retention time,SRT)为20 d,共稳定运行了3个SRT周期。稳定运行期间,以甲醇为碳源的SBR中MLSS保持在2 190~2 410 mg·L?1。每个SBR循环周期完成进水后,用0.1 mol·L?1盐酸调节SBR内的pH为7.0,实验期间室温为25~28 ℃。
1.2. 分析方法
1)各指标的测定。化学需氧量(chemical oxygen demand,COD)采用AQ4001测定仪(美国热电公司)测定,TN、NO3?-N、NO2?-N、NH4+-N和五日生化需氧量(biochemical oxygen demand after five days,BOD5)均采用标准方法[10]测定,pH和电导率采用835A奥立龙水质分析仪(美国热电公司)测定。2) DNA提取与PCR纯化。将SBR污泥样品200 g进行离心15 min,收集底部沉淀物抽提DNA。采用二轮PCR扩增和纯化。Illumina Miseq文库构建和测序:采用cutadapt软件检测接头和修剪末端,使用QIIME软件筛选序列和嵌合体,嵌合体的去除使用Usearch算法,操作分类单元(operational taxonomic unit,OTU)聚类算法使用Uclust。微生物群落多样性分析:将序列中相似度阈值为97%的归为OTU划分,采用RDP Classifier在80%置信度水平上进行物种分类。
3) qPCR定量。从SBR污泥中提取所需核酸物质,用Qubit? dsDNA HS Assay Kit测定纯化后的DNA浓度;PCR扩增反应体系为20 μL,荧光PCR数据采用数据软件进行分析。
2.1. 污染物去除及反硝化脱氮效果
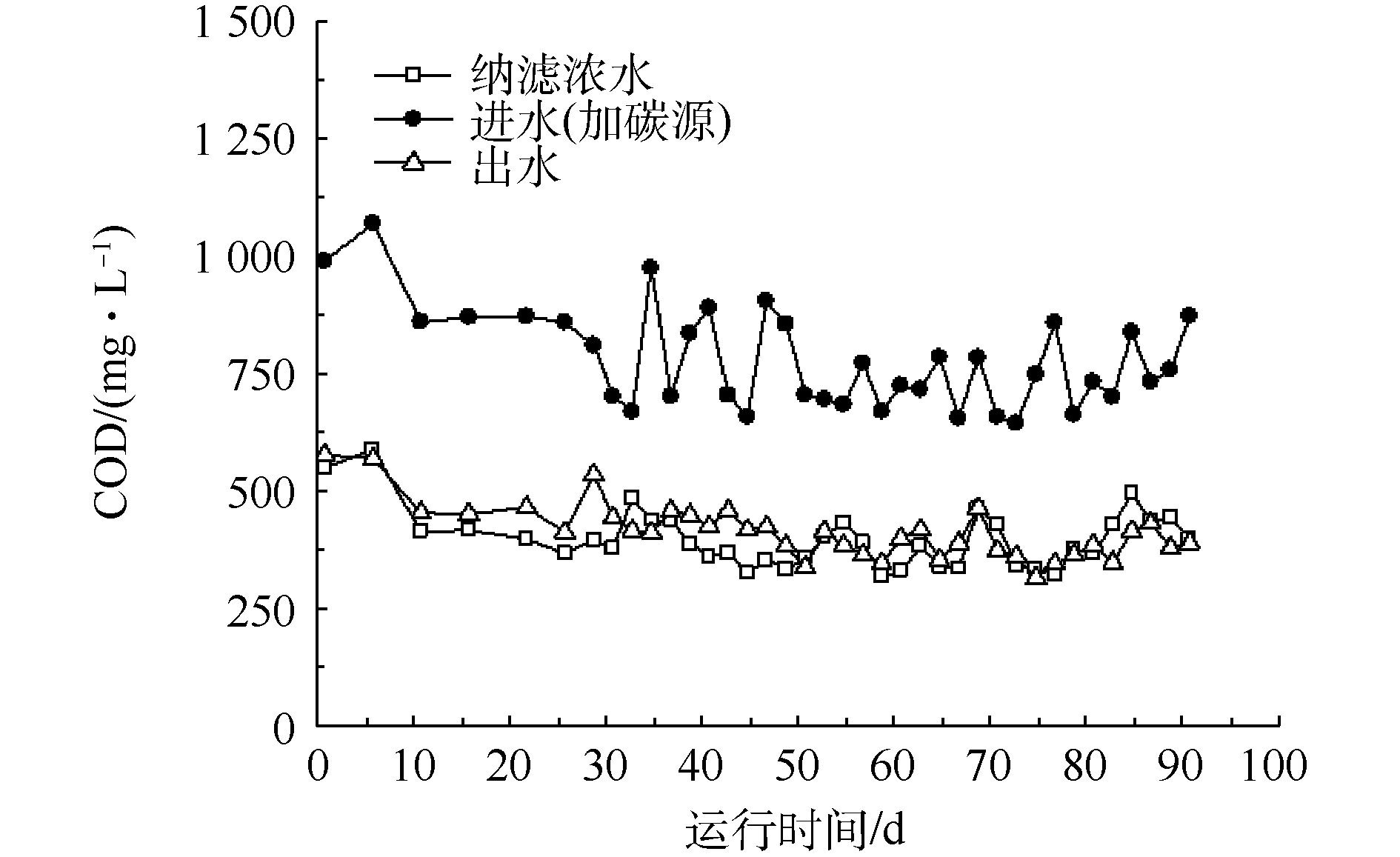
焦化纳滤浓水为难降解高盐废水,电导率为9 450~11 250 μS·cm?1、BOD5/COD低、可生化性差。有研究[11]表明,将NO3?-N转变为N2需要碳源作为还原反应的电子供体。在焦化纳滤浓水环境中,微生物生长繁殖和代谢可能受到盐分的抑制,为确保脱氮效率和反硝化完全,按照碳氮比8∶1投加甲醇。SBR共运行90 d,其中驯化30 d,稳定运行60 d。在SBR运行期间,进出水COD的变化如图1所示。在稳定运行期间,纳滤浓水的COD为318~496 mg·L?1;在添加碳源的SBR进水中,COD为617~975 mg·L?1;出水COD为328~474 mg·L?1。这说明加入的甲醇大部分被微生物降解了,浓水中的难降解有机物几乎没有被去除。
在驯化和稳定运行期间,焦化纳滤浓水SBR进出水中TN和NO3?-N的变化如图2所示。在稳定运行期间,纳滤浓水进水中TN和NO3?-N的平均浓度分别为58.9 mg·L?1和31.7 mg·L?1,而出水中两者平均浓度分别为24.6 mg·L?1和2.0 mg·L?1。系统对TN和NO3?-N的平均去除率分别为58.2%和93.8%。这说明在SBR系统中,甲醇作为电子供体时,反硝化反应完全。张玉莹等[12]发现,煤制气废水在缺氧条件下硝态氮的平均去除率为90%,本研究结果略高于其研究结果。在稳定运行期间,SBR出水亚硝态氮浓度为0.14~0.85 mg·L?1,说明焦化纳滤浓水在碳源充足的条件下,在微生物的反硝化过程中没有发生亚硝酸盐累积现象。
2.2. 微生物多样性分析
为研究以甲醇为碳源的焦化纳滤浓水在反硝化过程中微生物群落变化规律,分别采集SBR中的接种污泥、驯化完成时的污泥以及稳定运行中第51、71和91天的污泥,取样时间依次记为1、31、51、71和91 d。表1为高通量测序获得的污泥样品多样性指数。所有样品中共获得268 884条测序序列,32 440个操作分类单元(OTU)。在16S的V1~V3区进行扩增和测序,每个样品的测序量均在40 000以上,OTU数量均在5 000个以上。在测序过程中,香农指数(Shannon)为7.7~8.8,菌种丰富度指数(Chao1)为16 664.3~24 112.2,测序深度指数(goods’coverage)为0.87~0.91,辛普森指数(Simpson)为0.96~0.98,说明样品测序过程中物种总数量多样性丰富,可满足测序要求。
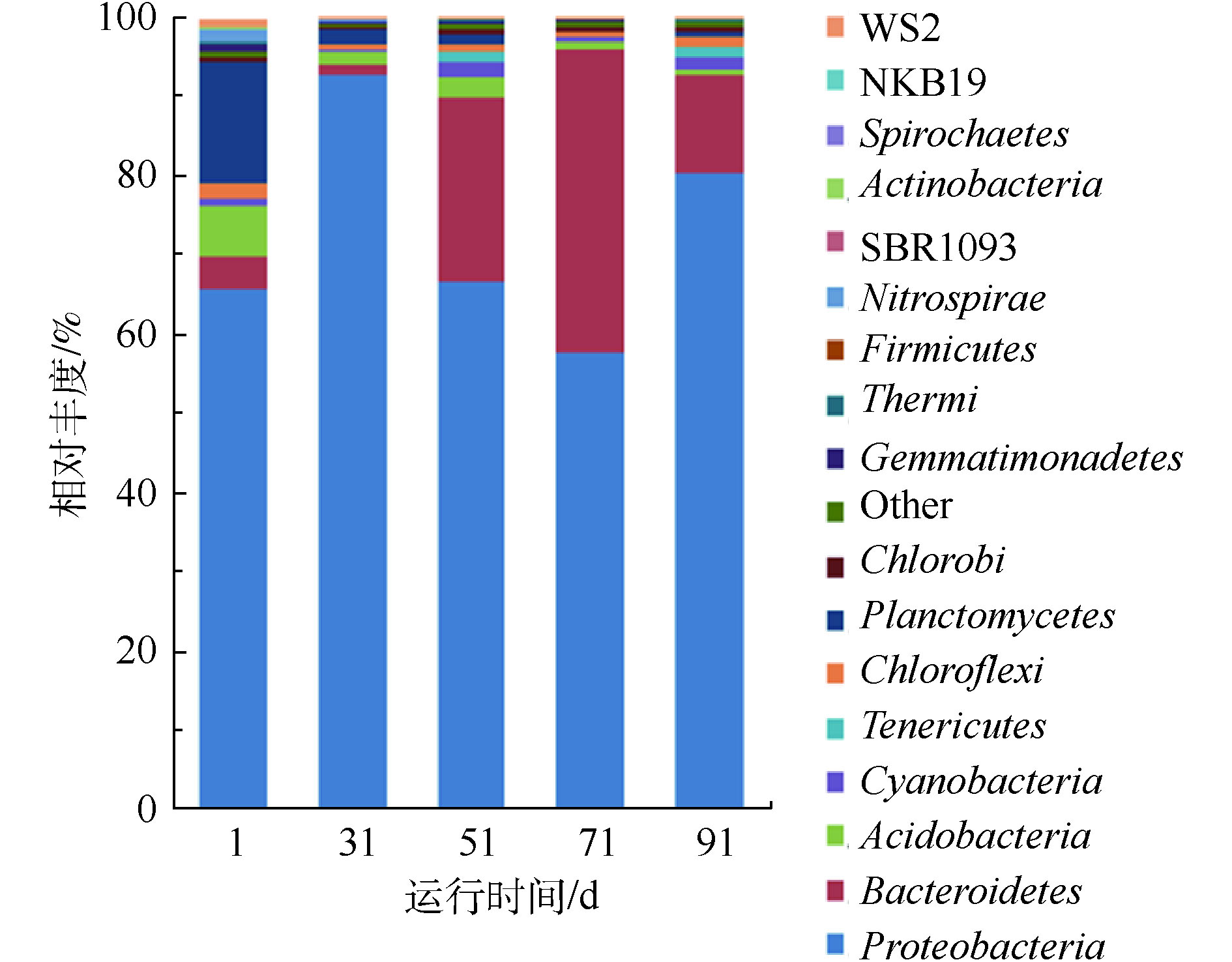
为分析SBR系统在处理纳滤浓水过程中微生物群落结构的变化,在门水平上对样品中微生物相对丰度进行了分析(见图3)。接种污泥(1 d)中微生物群落主要归属为变形菌门(相对丰度为65.2%)、浮霉菌门(相对丰度为15.7%)、酸杆菌门(相对丰度为6.7%)和拟杆菌门(相对丰度为3.9%)。经过30 d的驯化后,第31天的污泥中变形菌门的相对丰度从65.2%上升为92.9%,成为绝对的优势菌门;而浮霉菌门、酸杆菌门和拟杆菌门的相对丰度均明显降低。这说明污泥在驯化过程中,随着盐分的提高,变形菌门逐渐适应了焦化纳滤浓水的高电导率环境,而其他菌门的微生物活性受到抑制,相对丰度均有所降低。
在60 d的稳定运行期间,变形菌门仍然为最主要的优势菌门,在51、71和91 d的样品中,变形菌门相对丰度分别为66.8%、57.8%和80.4%。变形菌门为微生物中最丰富的菌门,包含多种生物代谢种类。已有研究表明,在焦化废水中,变形菌门能够完成降解有机物和生物脱氮的任务[13]。MA等[14]分析不同地域多套焦化废水生化系统菌群结构时发现,污泥中主要菌群均为变形菌门,平均相对丰度为76.1%。LORENZO等[15]发现,在多组高盐废水的生化实验中,变形菌门均为核心菌门,这说明变形菌门能适应高电导率的环境。在纳滤浓水的高电导率环境中,拟杆菌门在接种污泥中的相对丰度较低。在稳定运行过程中,拟杆菌门71 d的相对丰度最高,为38.2%;51 d和91 d时相对丰度分别为23.2%和12.5%,在纳滤浓水反硝化过程中逐渐成为了第2大菌门。拟杆菌门常见于工业废水生化系统中,可降解废水中蛋白质和糖类等有机化合物。ZHU等[16]指出,含盐量为2.5%的浓水生化系统可保持稳定的反硝化效率,通过菌群分析发现变形菌门和拟杆菌门为核心菌门。在纳滤浓水生化系统稳定运行期间,变形菌门和拟杆菌门的相对丰度之和为90.0%~96.0%。酸杆菌门和浮霉菌门为系统中的稀有菌门,相对丰度均低于2.7%。
在SBR运行过程中,污泥在纲层面上各分组中的丰度如图4所示。接种污泥中最主要的菌纲为β-变形菌纲(相对丰度为46.0%),其后的菌纲依次为浮霉菌纲(相对丰度为15.1%)、α-变形菌纲(相对丰度为11.5%)、chloracidobacteria(相对丰度为6.4%)、γ-变形菌纲(相对丰度为6.0%)、厌氧绳菌纲(相对丰度为1.7%)。驯化完成后,β-变形菌纲和α-变形菌纲相对丰度均提高,其他的菌纲相对丰度均减少。这说明在纳滤浓水30 d的驯化过程中,微生物系统中菌纲和相对丰度均发生了明显变化。
在稳定运行期间,在纲层面,β-变形菌纲为核心菌纲,相对丰度为38.7%~49.4%。有研究[17-18]指出,在废水的反硝化过程中,β-变形菌纲为核心菌纲,因为陶厄氏菌属和丛毛单胞菌属等重要反硝化功能菌属均属于β-变形菌纲。在稳定运行期间,α-变形菌纲相对丰度为12.5%~30.0%,γ-变形菌纲相对丰度为4.4%~10.8%。据报道[19-20],α-变形菌纲和γ-变形菌纲均适应高盐的工业废水环境,在缺氧条件下,α-变形菌纲可利用短链脂肪酸进行生物脱氮。在焦化纳滤浓水中运行过程中发现,变形菌门中不同菌纲的分布规律为β-变形菌纲>α-变形菌纲>γ-变形菌纲,这一规律和多套焦化废水生化系统中变形菌纲相对分布排列的研究结果[13]一致。在纳滤浓水反硝化过程中,拟杆菌门的黄杆菌纲也是重要的菌纲,稳定运行期间的相对丰度为8.3%~22.9%,这说明黄杆菌纲可适应高盐的浓水环境。O'REILLY等[21]发现,在海水沉积物中,黄杆菌纲为主要菌纲。另外,在稳定运行期间,纤维黏网菌纲为0.6%~8.7%,拟杆菌纲相对丰度为1.5%~4.8%。
2.3. 不同反硝化功能基因分析
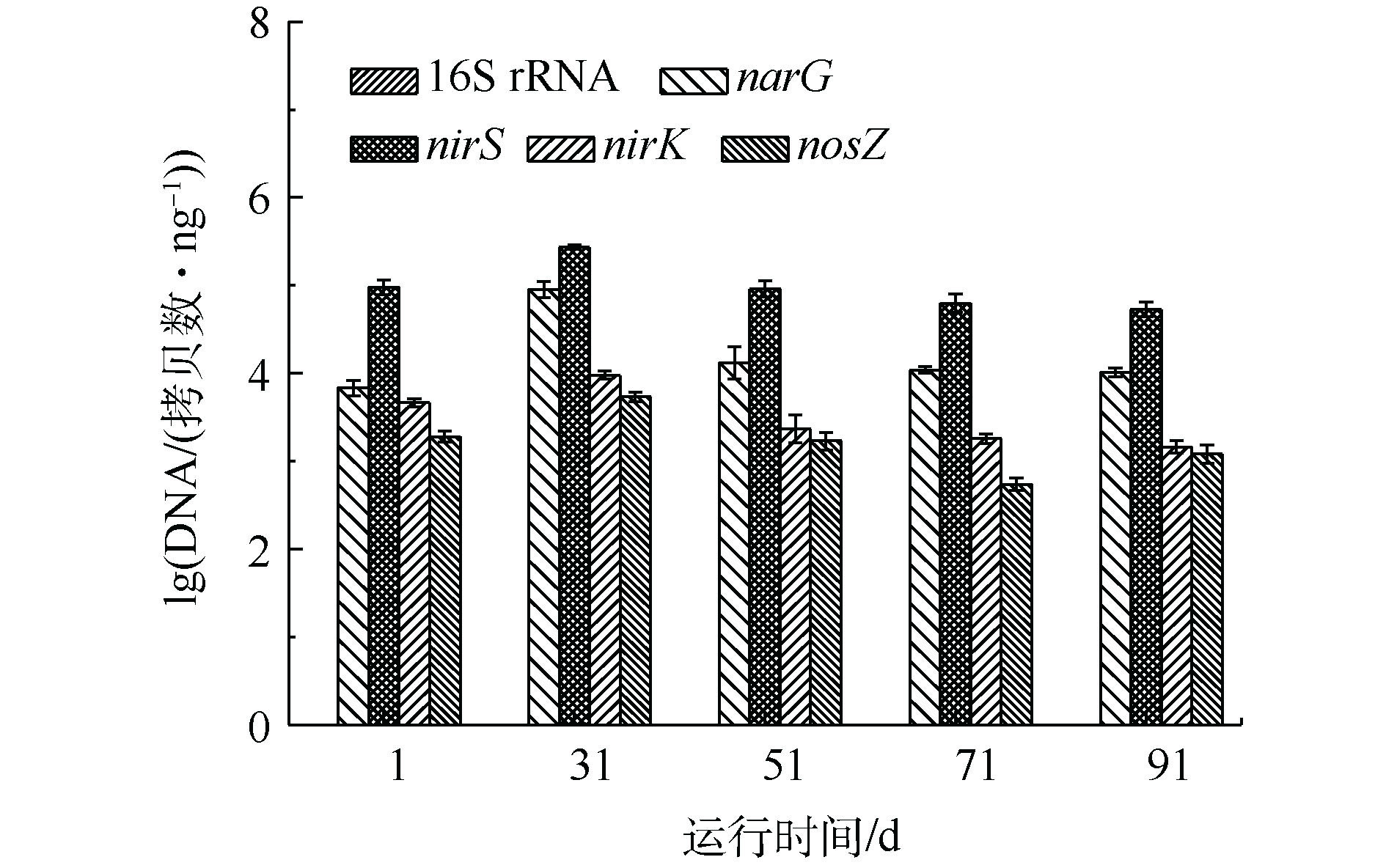
焦化纳滤浓水生物脱氮过程中,污泥中16S rRNA(细菌总数)、硝酸盐还原酶(narG)、亚硝酸盐还原酶(nirS和nirK)、氧化亚氮还原酶(nosZ)的绝对定量数据(qPCR)见图5。由图5可知,在SBR运行过程中,16S rRNA的拷贝数均高于接种污泥,这说明在碳源充足的条件下,适应焦化纳滤浓水的细菌总数保持稳定。narG可将细胞色素cdl-型亚硝酸盐还原酶nirS和Cu-型亚硝酸盐还原酶nirK均可将液态的NO2?-N转化为气态的一氧化氮,这是反硝化过程中关键的一步,它决定了反硝化的速率[23]。在纳滤浓水反硝化过程中,nirS为5.33×104~2.71×105拷贝数·ng?1,nirK为1.45×103~9.49×103拷贝数·ng?1。以上实验结果表明,在纳滤浓水反硝化过程中,nirS的拷贝数高于nirK约1~2个数量级。SHU等[24]深入研究了12个工业废水反硝化生化系统,发现nirS的拷贝数均远大于nirK。虽然nirS和nirK在生物脱氮过程功能和作用相同,可是含有nirK的微生物菌属偏向好氧环境,含有nirS的微生物菌属在兼氧和缺氧环境中相对丰度更多[25]。
NosZ可将氧化亚氮转化为氮气,完成了反硝化过程的最后一步。在稳定运行期间,nosZ为5.43×102~1.20×103拷贝数·ng?1;在纳滤浓水反硝化过程中,与narG、nirS和nirK相比,nosZ的拷贝数量最低,这是由于nosZ对电子供体的竞争能力最弱[26]。
在甲醇为碳源的焦化纳滤浓水反硝化过程中,出水硝态氮平均值为2.0 mg·L?1,narG、 nirS、nirK和nosZ等反硝化功能基因的数量达到了动态平衡,可确保
2)采集5个SBR污泥样品,采用高通量测序,共获得268 884条测序序列,32 440个操作分类单元。在稳定运行期间,变形菌门和拟杆菌门为核心菌门,两者的相对丰度之和为90.0%~96.0%;在菌纲层面,β-变形菌纲、α-变形菌纲和黄杆菌纲为主要菌纲。
3)焦化纳滤浓水反硝化运行中narG、nirS、nirK和nosZ等反硝化功能基因的数量均达到了动态平衡,nirS的拷贝数远高于nirK,说明nirS在亚硝酸转化过程中起到重要作用。
参考文献
 下载:
下载: 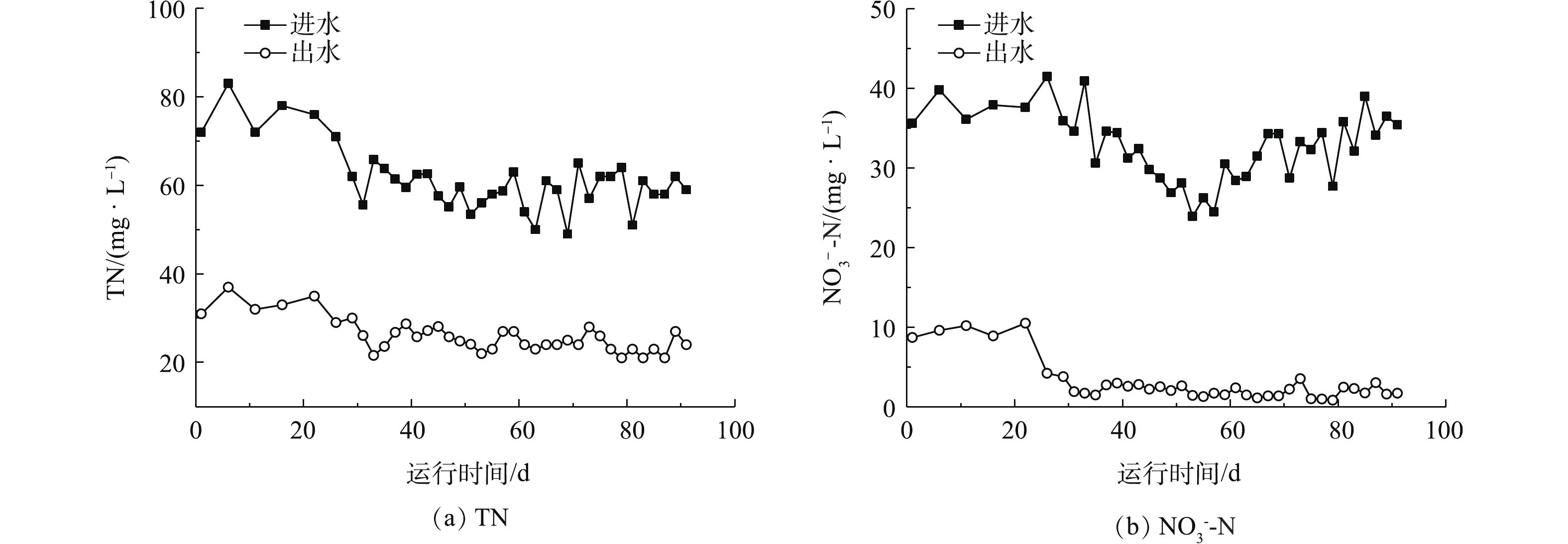

 点击查看大图
点击查看大图