 ,1, 喻树迅
,1, 喻树迅 ,2
,2Cloning and Drought Resistance Analysis of GhWRKY33 in Upland Cotton
WEI Xin1, WANG HanTao2, WEI HengLing2, FU XiaoKang2, MA Liang2, LU JianHua2, WANG XingFen ,1, YU ShuXun
,1, YU ShuXun ,2
,2通讯作者:
责任编辑: 李莉
收稿日期:2020-06-10接受日期:2020-06-28网络出版日期:2020-11-16
Received:2020-06-10Accepted:2020-06-28Online:2020-11-16
作者简介 About authors
魏鑫,Tel:17629552778;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (7104KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
魏鑫, 王寒涛, 魏恒玲, 付小康, 马亮, 芦建华, 王省芬, 喻树迅. 陆地棉GhWRKY33的克隆及抗旱功能分析[J]. 中国农业科学, 2020, 53(22): 4537-4549 doi:10.3864/j.issn.0578-1752.2020.22.002
WEI Xin, WANG HanTao, WEI HengLing, FU XiaoKang, MA Liang, LU JianHua, WANG XingFen, YU ShuXun.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】棉花是重要的经济作物,经常遭受干旱、盐碱等恶劣环境条件的影响,造成棉花产量和品质下降。当植物受到干旱胁迫后,会启动一系列干旱胁迫相关基因的表达,从分子水平、细胞水平以及生理生化水平做出相应的应激反应,从而实现对干旱胁迫的抗逆反应[1,2]。转录因子通常作为一类调控因子参与植物的生长发育及干旱等各种胁迫应答过程,因此对转录因子的功能进行研究具有重要意义[3]。【前人研究进展】WRKY转录因子是植物中最大的转录因子家族之一,由1个或2个WRKY保守结构域构成,N端是一段高度保守的序列(WRKYGQK),由大约60个保守的氨基酸残基组成,而C端是锌指结构(CX4-5CX22-23HX1H或CX7CX23HX1C)[4,5]。根据保守的WRKY结构域数量和锌指序列不同的特点,可将WRKY蛋白归为3组。Ⅰ组有2个WRKY结构域,锌指结构类型为C2H2,Ⅱ组和Ⅲ组包含1个WRKY结构域,锌指结构类型分别为C2H2和C2HC。根据WRKY结构域的进化关系和某些氨基酸基序,Ⅱ组又可以分为5个亚组:Ⅱa、Ⅱb、Ⅱc、Ⅱd和Ⅱe[6,7,8]。前人研究表明,许多WRKY转录因子在植物的生长发育、各种胁迫反应中具有重要作用[9,10,11,12],尤其在干旱胁迫中充当着十分重要的角色。CHU等[13]通过将陆地棉GhWRKY41转化烟草,发现该基因过量表达提高了转基因烟草的抗旱性水平,且干旱条件下植株的丙二醛含量降低,抗氧化酶活性增强,促进了气孔的关闭。烟草中过表达GhWRKY25,干旱条件下丙二醛、活性氧含量升高,超氧化物歧化酶、过氧化物酶、过氧化氢酶活性降低,植株抗旱能力下降[14]。在拟南芥中,AtWRKY44通过糖信号通路调控其对干旱胁迫的响应[15];过表达AtWRKY57提高了拟南芥的抗旱性和耐盐性[16];AtWRKY46被证实调节干旱和盐胁迫,且参与调节光依赖性的气孔开放[17]。MtWRKY76的过表达显著提高了转基因植株的抗旱性[18]。在葡萄中,VIWRKY48在干旱胁迫下也被证明能够提高过氧化氢酶、过氧化物酶、过氧化物歧化酶的抗氧化酶活性,不仅增加了葡萄的抗旱性,同时在抗白粉病中也发挥着重要作用[19]。【本研究切入点】棉花是非常重要的经济作物,但其产量和品质严重受到干旱、盐碱的影响。WRKY转录因子作为植物中最大的转录因子家族之一,与众多的非生物胁迫相关,但在棉花中与干旱相关的研究较少。【拟解决的关键问题】本研究通过克隆棉花GhWRKY33,并进行亚细胞定位,分析其在干旱及不同激素处理下的表达模式,在拟南芥中过量表达,明确其在抗旱中的作用,为棉花抗旱机制解析及分子育种奠定基础。1 材料与方法
1.1 材料
棉花材料中棉所10号以及拟南芥哥伦比亚野生型(Col-0生态型)均由中国农业科学院棉花研究所早熟课题组提供。
高保真酶、限制性内切酶、连接酶、质粒DNA提取试剂盒购自南京诺唯赞生物公司;PCR片段纯化试剂盒购自美基生物;植物RNA提取试剂盒购自北京天根生物公司;反转录试剂盒购自TaKaRa公司;荧光定量试剂购自康为世纪;试验转化用到的大肠杆菌,农杆菌感受态细胞均购自全式金公司;脯氨酸、丙二醛含量检测试剂盒购自于北京索莱宝科技有限公司;各种载体由中国农业科学院棉花研究所早熟课题组保存并提供;引物(表1)合成及测序工作由河南尚亚生物技术有限公司完成。
Table 1
表1
表1研究所用引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence(5′-3′) |
|---|---|
| GhWRKY33-F | CACGGGGGACTCTAGAATGGCTGCTTCATCATCATCT |
| GhWRKY33-R | GACGGCCAGTGAATTCTCAAGACAGGAATCCGTCCAA |
| qRT-GhWRKY33-F | GGGAAACCCCAATCCAAGGAGC |
| qRT-GhWRKY33-R | AATCATGGGATGCTCGCTCCAC |
| qRT-GhActin-F | ATCCTCCGTCTTGACCTTG |
| qRT-GhActin-R | TGTCCGTCAGGCAACTCAT |
| GhWRKY33-GFP-F | CACGGGGGACTCTAGAATGGCTGCTTCATCATCATCT |
| GhWRKY33-GFP-R | CTTTACTCATACTAGTAGACAGGAATCCGTCCAAAAATG |
| qRT-AtRD29A-F | ATCACTTGGCTCCACTGTTGTTC |
| qRT-AtRD29A-R | AAAACACACATAAACATCCAAAGT |
| qRT-AtCOR15A-F | ACTCAGTTCGTCGTCGTTTCT |
| qRT-AtCOR15A-R | CTTCTTTTCCTTTCTCCTCCAC |
| qRT-AtP5CS-F | GAGGGGGTATGACTGCAAAA |
| qRT-AtP5CS-R | AACAGGAACGCCACCATAAG |
| qRT-AtUBQ1-F | TGAGCCTTCCTTGATGATGCT |
| qRT-AtUBQ1-R | GCACTTGCGGCAAATCATCT |
新窗口打开|下载CSV
1.2 GhWRKY33的克隆
通过在线网站CottonFGD(1.3 生物信息学分析
使用SOPMA分析蛋白的二级结构;使用ProtScale(1.4 GhWRKY33的亚细胞定位
利用MEGA 6.0设计引物GhWRKY33-GFP-F/R,以pBI121-GhWRKY33过表达载体质粒为模板进行扩增,用限制性内切酶XbaⅠ、SpeⅠ对pBI121-GFP载体进行双酶切,利用同源重组法构建融合蛋白瞬时表达载体35S::GhWRKY33-GFP,通过农杆菌渗透法将融合表达载体注射到本氏烟草叶片中,同时注射pBI121-GFP载体作为对照,暗培养24 h后再在正常条件下培养24 h,利用激光共聚焦显微镜观察荧光信号。1.5 GhWRKY33的组织表达模式分析
为了研究GhWRKY33的组织表达模式,在开花期取中棉所10号的根、茎、叶片、雄蕊、雌蕊、顶芽、花瓣、萼片等组织,利用多糖多酚植物RNA提取试剂盒提取各组织的RNA,反转录合成cDNA,使用ABI7500 Real Time PCR Systerm进行实时荧光定量PCR试验,反应体系为2×Ultra SYBR Mixture 10 μL、Primer F 0.4 μL、Primer R 0.4 μL、cDNA 0.8 μL、ddH2O 8.4 μL。反应程序为95℃ 10 min;95℃ 15 s,60℃ 30 s,72℃ 32 s;95℃ 15 s,60℃ 1 min;95℃ 30 s,60℃ 15 s,30个循环。每种组织取3个生物学重复,并进行3次独立的试验,使用相对定量法2-ΔΔCt[20]进行数据分析。1.6 不同激素处理和干旱胁迫下的基因表达分析
将中棉所10号种植于温室,正常生长至3叶期,对棉苗进行激素和干旱处理。激素处理:分别取茉莉酸(100 μmol·L-1)、脱落酸(100 μmol·L-1)、乙烯(0.2%ET)溶液各10 mL,均匀喷洒至叶片表面;干旱处理:用自来水将棉苗的根冲洗干净后,将棉苗放置于锥形瓶中,加入18%的PEG6000溶液100 mL模拟干旱。每个处理选取5株棉苗,在处理后0、3、6、12、24和48 h时取样,用于基因表达分析,qRT-PCR反应体系和条件同1.4所述。1.7 转基因拟南芥的获得
将pBI121载体线性化,其酶切位点上游为XbaⅠ、下游为SacⅠ,利用同源重组的方法将目的片段与载体连接,并转化大肠杆菌DH5α,通过菌液PCR验证为阳性菌落后,送河南尚亚生物技术公司测序,从而获得序列正确的GhWRKY33片段。通过农杆菌介导的蘸花法转化拟南芥。将侵染后的拟南芥暗培养24 h,然后转移至正常条件培养室培养,一个月左右收获拟南芥种子,将种子30℃烘干2 d,用1% NaClO、75%的乙醇消毒,灭菌ddH2O清洗5次以上,平铺至含有卡那霉素的1/2MS培养板,4℃春化2 d,放置22℃、16 h光照/8 h黑暗的培养箱中培养筛选阳性植株,之后单株收获,以同样的方法筛选直至获得T3代纯合株系。
1.8 转基因拟南芥的抗旱性鉴定
1.8.1 干旱处理后拟南芥性状鉴定 分别取少量的Col-0野生型、3个转基因拟南芥株系OE-1、OE-2、OE-3种子洒在营养土中,一周之后挑选生长状态一致的幼苗移栽至营养钵中,每个株系移栽3盆,每盆4棵苗,作为重复,正常生长2周左右,使用20%的PEG6000溶液浇灌根系,每盆大约150 mL,处理12 h后观察野生型Col-0、转基因株系OE-1、OE-2、OE-3的萎蔫情况。1.8.2 脯氨酸含量测定 抗旱性强的植株会积累较多的脯氨酸,可以作为抗旱鉴定的指标之一。取干旱处理的野生型和3个转基因株系的拟南芥叶片约0.1 g,对照组和试验组均取3个生物学重复,按照试剂盒所述步骤操作:加入1 mL磺基水杨酸冰浴匀浆,沸水浴震荡10 min,10 000 r/min常温离心10 min,取上清静置冷却,取0.5 mL上清加入冰乙酸,0.5 mL脯氨酸提取液,沸水浴30 min,10 min震荡一次;冷却后加入1 mL甲苯震荡30 s,静置片刻,使色素转移至甲苯,于520 nm波长处比色,记录吸光值。标准曲线的制作:取试剂盒中标准品1 mg,分别配制为15、10、8、6、4、2、1和0 μg·mL-1,其余操作同样品的处理方法。
1.8.3 丙二醛含量测定 丙二醛作为质膜过氧化的产物之一,与植物耐旱性呈负相关,也可作为耐旱性指标之一。取野生型和3个转基因株系的拟南芥叶片0.1 g,每组样品取3个重复,按照试剂盒所述步骤操作:将组织放到研钵中加入1 mL提取液冰浴匀浆,8 000 r/min 4℃离心10 min,取上清置冰上待测;测定管中依次加入MDA检测工作液300 μL、样本100 μL、试剂三100 μL,空白管依次加入MDA监测工作液300 μL、蒸馏水100 μL、试剂三100 μL;混合液在100℃水浴保温60 min,冰浴冷却,10 000 r/min常温离心10 min,取200 μL上清,测定各样品在450、532和600 nm处吸光度,计算ΔA450=A450测定-A450空白、ΔA532=A532测定-A532空白、ΔA600=A600测定-A600空白。最终含量计算公式:MDA(nmol·mg-1)=5[6.45×(ΔA532- ΔA600)-0.56×ΔA450]/W。
2 结果
2.1 GhWRKY33的克隆与生物信息学分析
提取陆地棉中棉所10号总RNA,反转录后的cDNA作为模板,进行PCR扩增,得到目的片段与预期大小一致,约1 500 bp(图1),纯化后转入大肠杆菌并提取质粒。测序后,目的基因ORF全长1 533 bp,编码510个氨基酸残基,与参考基因组TM-1编号为Gh_D04G1318[21]的序列相似性达100%。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1GhWRKY33的PCR扩增
Fig. 1PCR amplification of GhWRKY33
SOPMA预测表明(图2-A),GhWRKY33蛋白的二级结构α螺旋(alpha helix)包含49个氨基酸残基,占9.61%;延伸链(extened strand)的氨基酸残基有47个,占9.22%;β-转角(beta turn)包含14个氨基酸残基,占2.75%;无规则卷曲(random coil)包含400个氨基酸残基,占78.43%,推测该蛋白的功能域主要由无规则卷曲构成。GhWRKY33蛋白含有26个苏氨酸(threonine)磷酸化位点(图2-B),可能与磷酸化调控有密切关系。亲疏水性分析显示(图2-C),亲水蛋白所占比例较大,属于亲水蛋白,其亲水性平均系数为-1.180。
图2
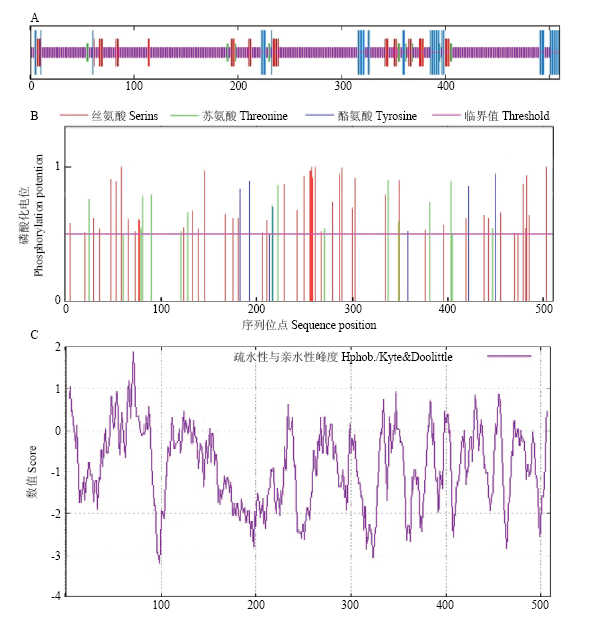 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2GhWRKY33编码蛋白的生物信息学分析
A:二级结构预测;B:磷酸化位点预测;C:亲疏水性分析(正峰代表疏水性,负峰代表亲水性)
Fig. 2Bioinformatic analysis of GhWRKY33
A: Secondary structure prediction; B: Phosphorylation site prediction; C: Hydrophobicity analysis (positive peak value represents hydrophobicity, negative peak value represents hydrophilicity)
使用PlantCARE软件对基因上游2 000 bp序列分析,发现存在多个顺式作用元件(表2)。如参与脱落酸调控反应的元件ABER、参与光反应的元件G-box、参与干旱胁迫响应的MYB结合位点、与植物组织分生表达相关的元件CAT-box等。
Table 2
表2
表2GhWRKY33启动子序列分析
Table 2
| 基序 Motif | 位置 Position (bp) | 序列 Sequence | 功能 Function |
|---|---|---|---|
| ABRE | 1252 | ACGTG | 参与脱落酸反应的顺式作用元件Cis-acting element involved in the abscisic acid responsiveness |
| ARE | 960 | AAACCA | 厌氧诱导所必需的顺式作用元件Cis-acting regulatory element essential for the anaerobic induction |
| 1858 | AAACCA | ||
| CAT-box | 1446 | GCCACT | 与分生组织表达相关的顺式作用元件Cis-acting regulatory element related to meristem expression |
| G-Box | 910 | CACGAC | 参与光响应的顺式作用元件Cis-acting regulatory element involved in light responsiveness |
| GCN4_motif | 222 | TGAGTCA | 胚乳表达中的顺式作用元件 Cis-regulatory element involved in endosperm expression |
| 1344 | TGAGTCA | ||
| 497 | TGAGTCA | ||
| MBS | 154 | CAACTG | 与干旱诱导相关的MYB结合位点MYB binding site involved in drought-inducibility |
新窗口打开|下载CSV
2.2 GhWRKY33蛋白同源比对及进化分析
利用NCBI在线数据库,选取多条相似度较高的WRKY蛋白与GhWRKY33进行比对,结果显示目的蛋白有2个WRKYGQK保守域,且锌指结构为C2H2,属于第Ⅰ类WRKY转录因子(图3)。进一步构建系统发育树(图4),结果显示,其与GrWRKY33同源关系最近,同源性达到了99%,其次是拟南芥等,而与大豆的同缘关系最远。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3GhWRKY33与其他物种WRKY蛋白序列比对分析
方框表示WRKY结构域,星号表示锌指结构。拟南芥AtWRKY33(NP_181381)、雷蒙德氏棉GrWRKY33(XP_012460140)、亚洲棉GaWRKY33(XP_017615740)、可可TcWRKY33(XP_017977471)、哥伦比亚锦葵HuWRKY33(XP_021290063)、榴莲DzWRKY33(XP_022767833)、黄麻CcWRKY33(OMO85052)、番木瓜CpWRKY33(XP_021908634)、橡胶树HbWRKY33(XP_021664002)、银白杨PaWRKY17(TKS07377)、木薯MeWRKY33(XP_021623467)、麻风树JcWRKY33(XP_012089749)、龙眼DlWRKY17(AEO31478)、橄榄树CaWRKY33(AXY96406)、山胡椒LgWRKY33(ALE71299)、豇豆VuWRKY33(QCE01025)、杨梅MrWRKY33(KAB1225722)、葡萄VvWRKY24(XP_002272040)、大豆GmWRKY49(NP_001304523)、大豆GmWRKY39(NP_001348302)、棉花GhWRKY33(XP_016680843)。下同
Fig. 3Sequence alignment of GhWRKY33 and WRKY proteins of other species The box indicates the WRKY domain, and the star indicates the zinc finger structure.
Arabidopsis thaliana AtWRKY33 (NP_181381), Gossypium raimondii GrWRKY33 (XP_012460140), Gossypium arboretum GaWRKY33 (XP_017615740), Theobroma cacao TcWRKY33 (XP_017977471), Herrania umbratica HuWRKY33 (XP_021290063), Durio zibethinus DzWRKY33 (XP_022767833), Corchorus capsularis CcWRKY33 (OMO85052), Carica papaya CpWRKY33 (XP_021908634), Hevea brasiliensis HbWRKY33 (XP_021664002), Populus alba PaWRKY17 (TKS07377), Manihot esculenta MeWRKY33 (XP_021623467), Jatropha curcas JcWRKY33 (XP_012089749), Dimocarpus longan DlWRKY17 (AEO31478), Canarium album CaWRKY33 (AXY96406), Lindera glauca LgWRKY33 (ALE71299), Vigna unguiculata VuWRKY33 (QCE01025), Morella rubra MrWRKY33 (KAB1225722), Vitis vinifera VvWRKY33 (XP_002272040), Glycine max GmWRKY49 (NP_001304523), Glycine max GmWRKY39 (NP_001348302), Gossypium hirsutum GhWRKY33 (XP_016680843). The same as below
图4
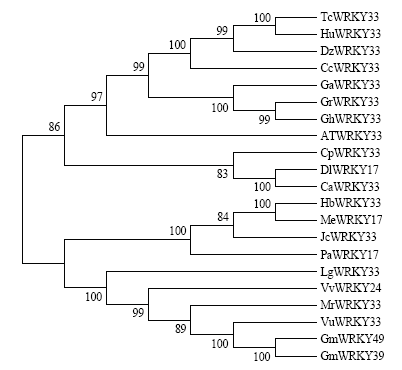 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4不同WRKY蛋白的进化树分析
Fig. 4Phylogenetic analysis of different WRKY proteins
2.3 GhWRKY33的组织特异性表达分析
以中棉所10号开花期的根、雄蕊、雌蕊、顶芽、叶片、茎、花瓣、萼片等组织为材料,对GhWRKY33进行组织特异性表达分析。结果显示,GhWRKY33具有明显的组织表达特异性,在芽中的表达量最高,其次是在雄蕊、雌蕊、叶片等组织,在根和茎中表达量最低(图5)。图5
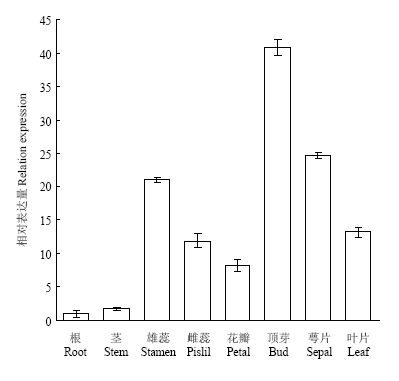 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5GhWRKY33组织特异性表达分析
Fig. 5Tissue specific expression analysis of GhWRKY33
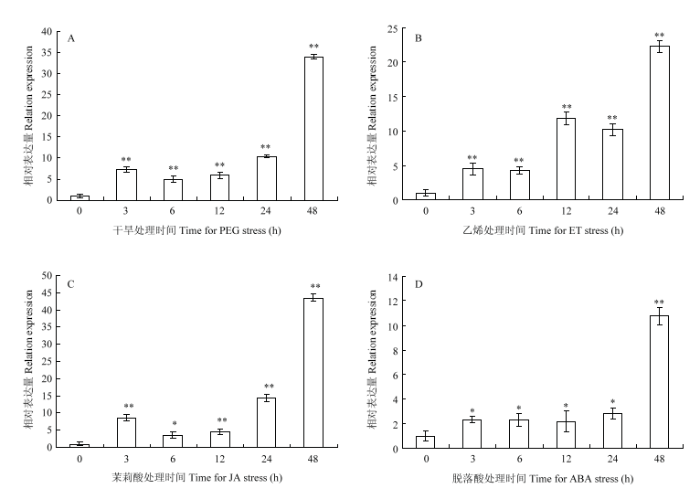
2.4 干旱和不同激素处理条件下基因的表达分析
qRT-PCR结果表明,该基因表达受到PEG、乙烯、茉莉酸、脱落酸的诱导。随着PEG、乙烯、茉莉酸处理时间的推移,该基因的表达量呈上升趋势,在48 h达到高峰(图6-A、图6-B和图6-C);脱落酸处理24 h内,基因的表达变化趋势不明显,但48 h时基因的表达量急剧上升,较之前的各时间点上升了10倍左右(图6-D)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同处理下GhWRKY33的表达模式
*和**:在P<0.05和P<0.01时差异达显著性和极显著性。下同
Fig. 6Expression patterns of GhWRKY33 under different treatments
* and **: Significant differences and significant differences at 0.05 level and 0.01 level. The same as below
2.5 GhWRKY33的亚细胞定位分析
以pBI121-GFP空载体为对照,将其与融合表达载体35S::GhWRKY33-GFP通过农杆菌介导分别转入本氏烟草。结果显示,对照组在细胞核和细胞膜上均出现绿色荧光信号,而融合蛋白35S::GhWRKY33- GFP只在细胞核中有绿色荧光信号(图7),表明该基因定位于细胞核中。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图735S::GhWRKY33-GFP融合蛋白的亚细胞定位
Fig. 7Subcellular localization of 35S::GhWRKY33-GFP fusion protein
2.6 转基因拟南芥的抗旱性鉴定及抗旱相关生理生 化指标分析
选取GhWRKY33表达量相对较高(图8)的3个T3代转基因拟南芥株系OE-1、OE-2、OE-3与野生型拟南芥同时浇灌等量的20% PEG6000溶液,发现12 h后野生型拟南芥和3个转基因株系都发生不同程度萎蔫,但是野生型较转基因株系萎蔫程度更加严重(图9),表明基因过量表达可以提高拟南芥的抗旱性水平。图8
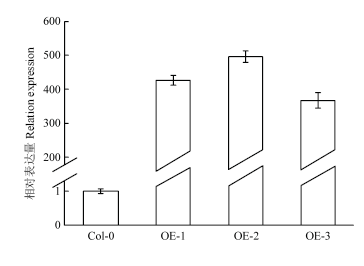 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8转基因拟南芥GhWRKY33的荧光定量表达分析
Fig. 8Quantitative expression level of GhWRKY33 gene in transgenic Arabidopsis
根据标准品的测定数据,建立脯氨酸的标准曲线(图10-A)。野生型拟南芥的吸光度为0.409,转基因拟南芥3个株系的吸光度分别为1.604、1.362和1.411,根据标准曲线得到野生型和3个转基因拟南芥株系的脯氨酸含量分别为3.843和26.875、22.062和22.997 μg·mL-1(图9-B),表明转基因株系的脯氨酸含量在干旱胁迫后极显著高于野生型。
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9PEG6000处理12 h野生型与转基因拟南芥生长状态
Fig. 9Growth of wild-type and transgenic Arabidopsis thaliana treated with PEG6000 for 12 h
图10
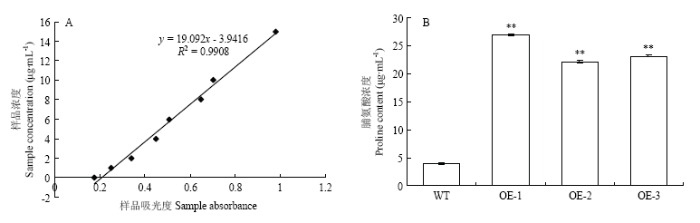 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10野生型与转基因拟南芥干旱处理12 h脯氨酸含量 A:标准曲线;B:野生型与转基因拟南芥脯氨酸的含量
Fig. 10Proline contents of wild-type and transgenic lines under drought treatment for 12 h A: Standard curve; B: The proline contents of wild type and of transgenic lines
丙二醛的测定结果表明,野生型拟南芥丙二醛含量为29.885 nmol·g-1,3个转基因株系中丙二醛含量为8.813、10.329和6.997 nmol·g-1,均极显著低于野生型(图11)。
图11
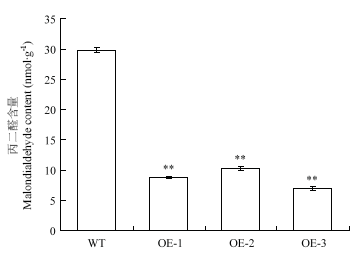 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图11野生型与转基因拟南芥干旱处理12 h丙二醛含量比较
Fig. 11MDA content of wild-type and transgenic lines under drought treatment for 12 h
2.7 GhWRKY33影响与干旱响应相关的基因表达
为了进一步研究GhWRKY33在干旱胁迫信号通路中的作用,鉴定了野生型和转基因拟南芥在PEG处理前后干旱响应基因AtRD29A、AtCOR15A、AtP5CS与GhWRKY33的表达水平(图12)。结果显示,正常生长条件下,野生型与转基因拟南芥中,AtRD29A、AtCOR15A和AtP5CS的表达水平没有明显差异,但在20%的PEG6000处理后,这些基因在转基因拟南芥中的表达量均极显著提高,表明植株受到干旱胁迫后,通过诱导GhWRKY33上调表达,进而正向调控干旱响应相关基因的表达,最终提高了转基因植株的抗旱性水平。图12
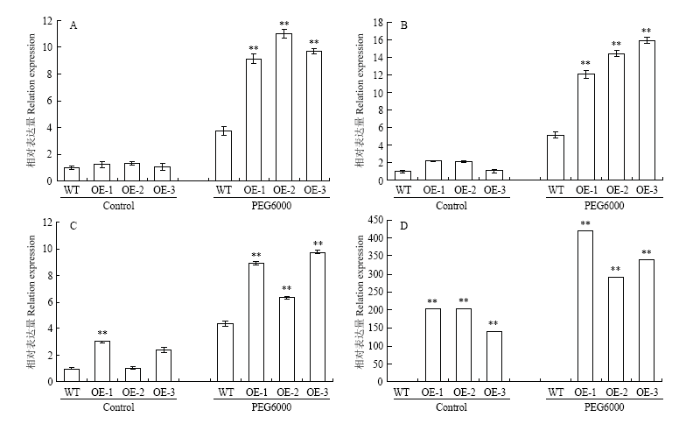 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图12野生型与转基因株系中干旱胁迫相关基因的表达
Fig. 12Expression analyses of the genes related to drought stress in the wild-type and transgenic lines
3 讨论
本研究从陆地棉中克隆了GhWRKY33,序列分析发现目的基因包含1 533 bp的开放阅读框,编码510个氨基酸残基,与其他WRKY蛋白序列比对发现该基因有2个WRKYGQK保守结构域,锌指蛋白为C2H2型,证明该基因属于第一类WRKY转录因子。在GhWRKY33中亲水蛋白占很大比重,使得该蛋白具有较强的亲水性,这种亲水性能够保持细胞的水分含量,也可以防止与其他蛋白脱水或凝聚[22]。在二级结构预测中,GhWRKY33蛋白中存在的α-螺旋,该结构的存在保障蛋白的柔韧性和流动性,在一定逆境下会发生结构变异[23],在受到干旱脱水影响时在一定程度上可以降低细胞所受伤害,这些蛋白结构的特性为该基因的抗旱性研究提供了一定的结构基础,亚细胞定位结果表明基因表达产物在细胞核内行使功能。通过对GhWRKY33的启动子区域的顺式元件分析,发现存在与干旱响应相关的MYB结合位点,并且在干旱处理下该基因的表达模式分析,证明其参与干旱胁迫相关的生物过程。
许多研究证明,WRKY转录因子受到外源激素的影响。陆地棉GhWRKY41可以通过ABA信号通路来调节植物的气孔关闭以及调节ROS的表达水平响应干旱和盐胁迫反应[13];GhWRKY17被ABA、H2O2、盐和干旱所诱导,在烟草中过量表达显著减低植株的耐盐性和耐旱性,证明了其通过ABA信号转导,调节植物中ROS的含量进而调控对盐和干旱的反应[24];烟草中,TaWRKY1依赖ABA途径调节气孔活动,保证水分的含量来抵御干旱,ABA受体基因NtPYL8在该信号通路中的传导作用至关重要[25];菊花中过表达CmWRKY1,与野生型相比,增加了植株对聚乙二醇的耐受性,并且初步证明是通过ABA信号通路响应干旱诱导[26];GmWRKY20在转基因拟南芥中增加了对ABA的敏感性,进而提高了对干旱胁迫的耐受性[27]。本研究分析了GhWRKY33在受到乙烯、茉莉酸和脱落酸诱导后的表达模式,结果表明,基因的表达量都呈现出显著的上调,推测该基因可能参与茉莉酸、乙烯以及脱落酸信号传导途径,进而参与调控植物的抗逆反应。
前人研究表明,1-吡咯啉-5-羧酸合成酶(P5CS)是参与脯氨酸合成的关键酶,而脯氨酸可以稳定原生质体胶体中的代谢过程并防止细胞脱水,通过增强P5CS的酶活性进而增加脯氨酸的生物合成,提高植物的抗逆性[28,29]。RD29A编码亲水蛋白,可以被ABA、干旱等胁迫诱导表达[30]。COR15A是干旱胁迫中的标志基因,通过ABA信号诱导参与干旱胁迫响应[31]。为了进一步了解GhWRKY33调控机制,本研究检测了干旱响应基因AtP5CS(AT2G39800)、AtRD29A(AT5G52310)、AtCOR15A(AT2G42540)在20%PEG6000处理前后的表达水平。发现这些基因在转基因拟南芥中的表达较野生型显著升高,因此,推测GhWRKY33通过ABA信号通路正向调控AtRD29A、AtCOR15A和AtP5CS的表达,从而提高了转基因拟南芥的抗旱性水平。关于GhWRKY33在棉花中的抗旱分子机制今后还需要进一步深入研究。
4 结论
从陆地棉中克隆获得GhWRKY33,属于典型的第Ⅰ类WRKY转录因子,其编码蛋白属于亲水蛋白;在顶芽中表达量最高,具有明显的组织表达特异性;GhWRKY33定位于细胞核;该基因响应干旱、乙烯、茉莉酸、脱落酸等的诱导。推测GhWRKY33作为正调控因子通过ABA信号通路调控拟南芥的抗旱性。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1093/jxb/erl164URLPMID:17075077 [本文引用: 1]

Plants respond to survive under water-deficit conditions via a series of physiological, cellular, and molecular processes culminating in stress tolerance. Many drought-inducible genes with various functions have been identified by molecular and genomic analyses in Arabidopsis, rice, and other plants, including a number of transcription factors that regulate stress-inducible gene expression. The products of stress-inducible genes function both in the initial stress response and in establishing plant stress tolerance. In this short review, recent progress resulting from analysis of gene expression during the drought-stress response in plants as well as in elucidating the functions of genes implicated in the stress response and/or stress tolerance are summarized. A description is also provided of how various genes involved in stress tolerance were applied in genetic engineering of dehydration stress tolerance in transgenic Arabidopsis plants.
DOI:10.1016/j.pbi.2007.04.014URL [本文引用: 1]

Plants must adapt to drought stress to survive. The phytohormone abscisic acid (ABA) is produced under drought stress conditions and is essential for the response to drought stress. The ABA level plays an important role in the response, and several enzymes for ABA biosynthesis and catabolism have been identified. Physiological studies have shown that several metabolites accumulate and function as osmolytes under drought stress conditions. Many drought-inducible genes with various functions have been identified, and transgenic plants that harbor these genes have shown increased tolerance to drought.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11103-008-9353-1URL [本文引用: 1]
[本文引用: 1]
DOI:10.13560/j.cnki.biotech.bull.1985.2016.10.009URL [本文引用: 1]

WRKY transcription factor families are characterized by a highly conserved WRKYGQK domain and involved in plant development,metabolism,answering to comprehensive biotic or abiotic stress. Recently,the research of WRKY transcription factors concentrate on stress response signaling network in different species. It reviewed progress of WRKYs members,and indicated that WRKY transcription factors play a heavy role in plant growth and regulating stress response. At the same time,there is less reported of WRKYs function in plant species besides model plant Arabidopsis thaliana and most of them focus on systematic research and analysis. In addition,numerous networks of WRKY transcription factors are still unclear.
DOI:10.13560/j.cnki.biotech.bull.1985.2016.10.009URL [本文引用: 1]

WRKY transcription factor families are characterized by a highly conserved WRKYGQK domain and involved in plant development,metabolism,answering to comprehensive biotic or abiotic stress. Recently,the research of WRKY transcription factors concentrate on stress response signaling network in different species. It reviewed progress of WRKYs members,and indicated that WRKY transcription factors play a heavy role in plant growth and regulating stress response. At the same time,there is less reported of WRKYs function in plant species besides model plant Arabidopsis thaliana and most of them focus on systematic research and analysis. In addition,numerous networks of WRKY transcription factors are still unclear.
DOI:10.1186/1471-2229-10-281URL [本文引用: 1]
DOI:10.1016/j.febslet.2005.01.057URLPMID:15733871 [本文引用: 1]

Calmodulin (CaM) is a ubiquitous Ca(2+)-binding protein known to regulate diverse cellular functions by modulating the activity of various target proteins. We isolated a cDNA encoding AtWRKY7, a novel CaM-binding transcription factor, from an Arabidopsis expression library with horseradish peroxidase-conjugated CaM. CaM binds specifically to the Ca(2+)-dependent CaM-binding domain (CaMBD) of AtWRKY7, as shown by site-directed mutagenesis, a gel mobility shift assay, a split-ubiquitin assay, and a competition assay using a Ca2+/CaM-dependent enzyme. Furthermore, we show that the CaMBD of AtWRKY7 is a conserved structural motif (C-motif) found in group IId of the WRKY protein family.
DOI:10.1007/s00425-011-1423-yURL [本文引用: 1]

WRKY proteins are a large super family of transcriptional regulators primarily involved in various plant physiological programs. In present study, the expression profile and putative function of the WRKY transcriptional factor, WRKY78, in rice were identified. Real-time RT-PCR analysis showed that OsWRKY78 transcript was most abundant in elongating stems though its expression was detected in all the tested organs. The expression profiles were further confirmed by using promoter-GUS analysis in transgenic rice. OsWRKY78::GFP fusion gene transient expression analysis demonstrated that OsWRKY78 targeted to the nuclei of onion epidermal cell. Furthermore, OsWRKY78 RNAi and overexpression transgenic rice lines were generated. Transgenic plants with OsWRKY78 overexpression exhibited a phenotype identical to the wild type, whereas inhibition of OsWRKY78 expression resulted in a semi-dwarf and small kernel phenotype due to reduced cell length in transgenic plants. In addition, a T-DNA insertion mutant line oswrky78 was identified and a phenotype similar to that of RNAi plants was also observed. Grain quality analysis data showed no significant differences, with the exception of minor changes in endosperm starch crystal structure in RNAi plants. Taken together, these results suggest that OsWRKY78 may acts as a stem elongation and seed development regulator in rice.
URL [本文引用: 1]

Partial sequences of a number of cDNAs showing homology with WRKY genes were amplified from two-day cold-acclimated Poncirus trifoliata by reverse transcription-polymerase chain reaction (RT-PCR) using degenerate primers designed based on conserved signature sequences of the WRKY domain. The full-length sequence of one of these cDNAs designated as PtrWRKY2 was obtained using rapid amplification of cDNA ends (RACE) method. PtrWRKY2 cDNA was 2,070 bp in length containing an open reading frame (ORF) encoding a polypeptide of 540 amino acids. The polypeptide was homologous to WRKY transcription factors (TFs) having double WRKY domains with Cys(2)His(2) signature motif. The PtrWRKY2 gene was clustered with putative WRKY TFs having double WRKY domains and showed close phylogenetic relation with putative WRKY TFs mostly from woody perennial plants. The expression of the PtrWRKY2 gene was analyzed by Northern blot in cold-hardy Poncirus and cold-sensitive Citrus species, Citrus grandis (pummelo) having the same gene with more than 95% nucleotide sequence homology. The results revealed that the expression of the PtrWRKY2 gene was initially induced in response to cold in both cold-hardy Poncirus and cold-sensitive pummelo; however, the expression was decreased at 1 h of cold acclimation and then, clearly repressed in Poncirus and pummelo. On the other hand, drought stress had no effect on the expression of the PtrWRKY2 gene in pummelo, but its expression was repressed by drought stress in Poncirus indicating a negative regulation. The results showed that PtrWRKY2 encodes a WRKY TF and its expression showed differential responses to cold acclimation and dehydration stress in Poncirus and Citrus.
DOI:10.1111/ppl.12651URLPMID:29027659 [本文引用: 1]

WRKY transcription factors are transcriptional regulators of signaling pathways involved in biotic and abiotic stress responses. In this study, we report that ectopic expression of the GhWRKY6-like gene significantly improved salt tolerance in Arabidopsis thaliana while silencing the GhWRKY6-like increase the sensitivity to abiotic stresses in cotton. GhWRKY6-like was localized to the nucleus. Expression of GhWRKY6-like was remarkably induced by salt, polyethylene glycol (PEG) and abscisic acid (ABA) treatments. For further characterization, the GhWRKY6-like gene was cloned and transformed into Arabidopsis. Our findings showed that the germination rate and root length were significantly improved in plants overexpressing GhWRKY6-like vs wild type (WT) under salt, mannitol and ABA treatments. Additionally, the overexpressing lines showed greater salt tolerance than WT plants in soil. In addition, overexpressing plants accumulated less H2 O2 and malondialdehyde (MDA), while higher proline content, superoxide dismutase (SOD) and peroxidase (POD) activities were detected under salt and osmotic stresses. In contrast, virus-induced gene silencing (VIGS) of GhWRKY6-like in cotton showed enhanced sensitivity compared to WT plants during salt and drought stresses. Additionally, expression analysis of stress-responsive genes in GhWRKY6-like Arabidopsis revealed that there was increased expression of genes involved in the ABA signaling pathway (AtABF4, AtABI5 and AtMYC2) and osmotic stress (AtSOS2, AtRD29a and AtRD29b). Our results revealed that GhWRKY6-like enhanced salt tolerance in Arabidopsis by scavenging reactive oxygen species and regulating the ABA signaling pathway. We suggest that overexpression of the GhWRKY6-like gene in cotton will enhance tolerance against salt, drought and osmotic stresses.
DOI:10.1016/j.plantsci.2012.08.013URL [本文引用: 1]

WRKY transcription factors regulate biotic, abiotic, and developmental processes. In terms of plant defense, WRKY factors have important roles as positive and negative regulators via transcriptional regulation or protein-protein interaction. Here, we report the characterization of the gene encoding Capsicum annuum WRKY transcription factor d (CaWRKYd) isolated from microarray analysis in the Tobacco mosaic virus (TMV)-P-0-inoculated hot pepper plants. CaWRKYd belongs to the WRKY Ila group, a very small clade in the WRKY subfamily, and WRKY ha group has positive/negative regulatory roles in Arabidopsis and rice. CaWRKYd transcripts were induced by various plant defense-related hormone treatments and TMV-P-0 inoculation. Silencing of CaWRKYd affected TMV-P-0-mediated hypersensitive response (HR) cell death and accumulation of TMV-P-0 coat protein in local and systemic leaves. Furthermore, expression of some pathogenesis-related (PR) genes and HR-related genes was reduced in the CaWRKYd-silenced plants compared with TRV2 vector control plants upon TMV-P-0 inoculation. CaWRKYd was confirmed to bind to the W-box. Thus CaWRKYd is a newly identified Capsicum annuum WRKY transcription factor that appears to be involved in TMV-P-0-mediated HR cell death by regulating downstream gene expression. (C) 2012 Elsevier Ireland Ltd.
DOI:10.1371/journal.pone.0143002URL [本文引用: 2]
DOI:10.1007/s00709-015-0885-3URLPMID:26410829 [本文引用: 1]

WRKY transcription factors are involved in various processes, ranging from plant growth to abiotic and biotic stress responses. Group I WRKY members have been rarely reported compared with group II or III members, particularly in cotton (Gossypium hirsutum). In this study, a group I WRKY gene, namely, GhWRKY25, was cloned from cotton and characterized. Expression analysis revealed that GhWRKY25 can be induced or deduced by the treatments of abiotic stresses and multiple defense-related signaling molecules. Overexpression of GhWRKY25 in Nicotiana benthamiana reduced plant tolerance to drought stress but enhanced tolerance to salt stress. Moreover, more MDA and ROS accumulated in transgenic plants after drought treatment with lower activities of SOD, POD, and CAT. Our study further demonstrated that GhWRKY25 overexpression in plants enhanced sensitivity to the fungal pathogen Botrytis cinerea by reducing the expression of SA or ET signaling related genes and inducing the expression of genes involved in the JA signaling pathway. These results indicated that GhWRKY25 plays negative or positive roles in response to abiotic stresses, and the reduced pathogen resistance may be related to the crosstalk of the SA and JA/ET signaling pathways.
DOI:10.1038/415977aURLPMID:11875555 [本文引用: 1]

There is remarkable conservation in the recognition of pathogen-associated molecular patterns (PAMPs) by innate immune responses of plants, insects and mammals. We developed an Arabidopsis thaliana leaf cell system based on the induction of early-defence gene transcription by flagellin, a highly conserved component of bacterial flagella that functions as a PAMP in plants and mammals. Here we identify a complete plant MAP kinase cascade (MEKK1, MKK4/MKK5 and MPK3/MPK6) and WRKY22/WRKY29 transcription factors that function downstream of the flagellin receptor FLS2, a leucine-rich-repeat (LRR) receptor kinase. Activation of this MAPK cascade confers resistance to both bacterial and fungal pathogens, suggesting that signalling events initiated by diverse pathogens converge into a conserved MAPK cascade.
DOI:10.3389/fpls.2016.00145URLPMID:26904091 [本文引用: 1]

Drought stress is a severe environmental factor that greatly restricts plant distribution and crop production. Recently, we have found that overexpressing AtWRKY57 enhanced drought tolerance in Arabidopsis thaliana. In this study, we further reported that the Arabidopsis WRKY57 transcription factor was able to confer drought tolerance to transgenic rice (Oryza sativa) plants. The enhanced drought tolerance of transgenic rice was resulted from the lower water loss rates, cell death, malondialdehyde contents and relative electrolyte leakage while a higher proline content and reactive oxygen species-scavenging enzyme activities was observed during stress conditions. Moreover, further investigation revealed that the expression levels of several stress-responsive genes were up-regulated in drought-tolerant transgenic rice plants, compared with those in wild-type plants. In addition to the drought tolerance, the AtWRKY57 over-expressing plants also had enhanced salt and PEG stress tolerances. Taken together, our study indicates that over-expressing AtWRKY57 in rice improved not only drought tolerance but also salt and PEG tolerance, demonstrating its potential role in crop improvement.
DOI:10.1111/tpj.12538URLPMID:24773321 [本文引用: 1]

Drought and salt stress severely inhibit plant growth and development; however, the regulatory mechanisms of plants in response to these stresses are not fully understood. Here we report that the expression of a WRKY transcription factor WRKY46 is rapidly induced by drought, salt and oxidative stresses. T-DNA insertion of WRKY46 leads to more sensitivity to drought and salt stress, whereas overexpression of WRKY46 (OV46) results in hypersensitivity in soil-grown plants, with a higher water loss rate, but with increased tolerance on the sealed agar plates. Stomatal closing in the OV46 line is insensitive to ABA because of a reduced accumulation of reactive oxygen species (ROS) in the guard cells. We further find that WRKY46 is expressed in guard cells, where its expression is not affected by dehydration, and is involved in light-dependent stomatal opening. Microarray analysis reveals that WRKY46 regulates a set of genes involved in cellular osmoprotection and redox homeostasis under dehydration stress, which is confirmed by ROS and malondialdehyde (MDA) levels in stressed seedlings. Moreover, WRKY46 modulates light-dependent starch metabolism in guard cells via regulating QUA-QUINE STARCH (QQS) gene expression. Taken together, we demonstrate that WRKY46 plays dual roles in regulating plant responses to drought and salt stress and light-dependent stomatal opening in guard cells.
[本文引用: 1]
DOI:10.1007/s11240-017-1335-zURL [本文引用: 1]
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
DOI:10.1038/nbt.3207URLPMID:25893781 [本文引用: 1]

Upland cotton is a model for polyploid crop domestication and transgenic improvement. Here we sequenced the allotetraploid Gossypium hirsutum L. acc. TM-1 genome by integrating whole-genome shotgun reads, bacterial artificial chromosome (BAC)-end sequences and genotype-by-sequencing genetic maps. We assembled and annotated 32,032 A-subgenome genes and 34,402 D-subgenome genes. Structural rearrangements, gene loss, disrupted genes and sequence divergence were more common in the A subgenome than in the D subgenome, suggesting asymmetric evolution. However, no genome-wide expression dominance was found between the subgenomes. Genomic signatures of selection and domestication are associated with positively selected genes (PSGs) for fiber improvement in the A subgenome and for stress tolerance in the D subgenome. This draft genome sequence provides a resource for engineering superior cotton lines.
DOI:10.1007/s00114-007-0254-yURLPMID:17479232 [本文引用: 1]

Research into late embryogenesis abundant (LEA) proteins has been ongoing for more than 20 years but, although there is a strong association of LEA proteins with abiotic stress tolerance particularly dehydration and cold stress, for most of that time, their function has been entirely obscure. After their initial discovery in plant seeds, three major groups (numbered 1, 2 and 3) of LEA proteins have been described in a range of different plants and plant tissues. Homologues of groups 1 and 3 proteins have also been found in bacteria and in certain invertebrates. In this review, we present some new data, survey the biochemistry, biophysics and bioinformatics of the LEA proteins and highlight several possible functions. These include roles as antioxidants and as membrane and protein stabilisers during water stress, either by direct interaction or by acting as molecular shields. Along with other hydrophilic proteins and compatible solutes, LEA proteins might also serve as
DOI:10.1002/bip.21693URLPMID:23325560 [本文引用: 1]

The occurrence of a highly conserved 11-mer repeating motif in the primary sequence is a major characteristic of group 3 late embryogenesis abundant (LEA3) proteins, which are strongly associated with abiotic stress tolerance of the plants. In this study, the three-dimensional structure, mimetic membrane association, and salt effect for consensus 11-mer motif from soybean PM2 protein (LEA3) were investigated in sodium dodecyl sulfate (SDS) micelles by NMR techniques. It was shown that the 11-mer motif was disordered in aqueous solution, but adopted an alpha-helix in SDS micelles. NMR diffusion measurements demonstrated that the 11-mer motif was associated with SDS micelles. Paramagnetic quenching NMR experiments further revealed the orientation of the 11-mer motif with respect to the mimetic membrane: the ordered N-terminal segment was inserted into the mimetic membrane, and the disordered C-terminal segment was exposed to water. In addition, salt addition could not change the secondary structure of the 11-mer motif, but might slightly alter the relative spatial position of some N-terminal residue atoms. These results implied that the 11-mer motif would take an important role in structural plasticity and membrane stabilization for LEA3 proteins.
DOI:10.1093/pcp/pcu133URLPMID:25261532 [本文引用: 1]

Drought and high salinity are two major environmental factors that significantly limit the productivity of agricultural crops worldwide. WRKY transcription factors play essential roles in the adaptation of plants to abiotic stresses. However, WRKY genes involved in drought and salt tolerance in cotton (Gossypium hirsutum) are largely unknown. Here, a group IId WRKY gene, GhWRKY17, was isolated and characterized. GhWRKY17 was found to be induced after exposure to drought, salt, H2O2 and ABA. The constitutive expression of GhWRKY17 in Nicotiana benthamiana remarkably reduced plant tolerance to drought and salt stress, as determined through physiological analyses of the germination rate, root growth, survival rate, leaf water loss and Chl content. GhWRKY17 transgenic plants were observed to be more sensitive to ABA-mediated seed germination and root growth. However, overexpressing GhWRKY17 in N. benthamiana impaired ABA-induced stomatal closure. Furthermore, we found that GhWRKY17 modulated the increased sensitivity of plants to drought by reducing the level of ABA, and transcript levels of ABA-inducible genes, including AREB, DREB, NCED, ERD and LEA, were clearly repressed under drought and salt stress conditions. Consistent with the accumulation of reactive oxygen species (ROS), reduced proline contents and enzyme activities, elevated electrolyte leakage and malondialdehyde, and lower expression of ROS-scavenging genes, including APX, CAT and SOD, the GhWRKY17 transgenic plants exhibited reduced tolerance to oxidative stress compared with wild-type plants. These results therefore indicate that GhWRKY17 responds to drought and salt stress through ABA signaling and the regulation of cellular ROS production in plants.
DOI:10.1007/s11105-016-0991-1URL [本文引用: 1]
DOI:10.1371/journal.pone.0150572URLPMID:26938878 [本文引用: 1]

WRKY transcription factors serve as antagonistic or synergistic regulators in a variety of abiotic stress responses in plants. Here, we show that CmWRKY1, a member of the group IIb WRKY family isolated from Chrysanthemum morifolium, exhibits no transcriptional activation in yeast cells. The subcellular localization examination showed that CmWRKY1 localizes to the nucleus in vivo. Furthermore, CmWRKY1-overexpressing transgenic lines exhibit enhanced dehydration tolerance in response to polyethylene glycol (PEG) treatment compared with wild-type plants. We further confirmed that the transgenic plants exhibit suppressed expression levels of genes negatively regulated by ABA, such as PP2C, ABI1 and ABI2, and activated expression levels of genes positively regulated by ABA, such as PYL2, SnRK2.2, ABF4, MYB2, RAB18, and DREB1A. Taken together, our results indicate that CmWRKY1 plays an important role in the response to drought in chrysanthemum through an ABA-mediated pathway.
DOI:10.1093/jxb/ert073URLPMID:23606412 [本文引用: 1]

The WRKY-type transcription factors are involved in plant development and stress responses, but how the regulation of stress tolerance is related to plant development is largely unknown. GsWRKY20 was initially identified as a stress response gene using large-scale Glycine soja microarrays. Quantitative reverse transcription-PCR (qRT-PCR) showed that the expression of this gene was induced by abscisic acid (ABA), salt, cold, and drought. Overexpression of GsWRKY20 in Arabidopsis resulted in a decreased sensitivity to ABA during seed germination and early seedling growth. However, compared with the wild type, GsWRKY20 overexpression lines were more sensitive to ABA in stomatal closure, and exhibited a greater tolerance to drought stress, a decreased water loss rate, and a decreased stomatal density. Moreover, microarray and qRT-PCR assays showed that GsWRKY20 mediated ABA signalling by promoting the expression of negative regulators of ABA signalling, such as AtWRKY40, ABI1, and ABI2, while repressing the expression of the positive regulators of ABA, for example ABI5, ABI4, and ABF4. Interestingly, GsWRKY20 also positively regulates the expression of a group of wax biosynthetic genes. Further, evidence is provided to support that GsWRKY20 overexpression lines have more epicuticular wax crystals and a much thicker cuticle, which contribute to less chlorophyll leaching compared with the wild type. Taken together, the findings reveal an important role for GsWRKY20 in enhancing drought tolerance and regulating ABA signalling.
DOI:10.1016/j.envexpbot.2005.12.006URL [本文引用: 1]
DOI:10.1073/pnas.89.19.9354URLPMID:1384052 [本文引用: 1]

Many plants synthesize and accumulate proline in response to osmotic stress. Despite the importance of this pathway, however, the exact metabolic route and enzymes involved in the synthesis of proline in plants have not been unequivocally identified. We report here the isolation of a mothbean (Vigna aconitifolia) cDNA clone encoding a bifunctional enzyme, delta 1-pyrroline-5-carboxylate synthetase (P5CS), with both gamma-glutamyl kinase and glutamic-gamma-semialdehyde dehydrogenase activities that catalyzes the first two steps in proline biosynthesis. The two enzymatic domains of P5CS correspond to the ProB and ProA proteins of Escherichia coli and contain a leucine zipper in each domain, which may facilitate inter- or intramolecular interaction of this protein. The Vigna P5CS enzyme activity is feedback regulated by proline but is less sensitive to end-product inhibition than is the E. coli gamma-glutamyl kinase. The P5CS gene is expressed at high levels in Vigna leaves and is inducible in roots subjected to salt stress, suggesting that P5CS plays a key role in proline biosynthesis, leading to osmoregulation in plants.
DOI:10.1007/s00425-011-1387-yURL [本文引用: 1]

Abiotic stresses have adverse effects on plant growth and productivity. The homologous RD29A and RD29B genes are exquisitely sensitive to various abiotic stressors. Therefore, RD29A and RD29B gene sequences have potential to confer abiotic stress resistance in crop species grown in arid and semi-arid regions. To our knowledge, no information on the physiological roles of the proteins encoded by RD29A and RD29B are available in the literature. To understand how these proteins function, we used reverse genetic approaches, including identifying rd29a and rd29b T-DNA knockout mutants, and examining the effects of complementing transgenes with the genes under control of their native promoters and chimeric genes with the native promoters swapped. Four binary vectors with the RD29A and RD29B promoters upstream of the cognate RD29A and RD29B cDNAs and as chimeric genes with noncognate promoters were used to transform rd29a and rd29b plants. Cold, drought, and salt induced both genes; the promoter of RD29A was found to be more responsive to drought and cold stresses, whereas the promoter of RD29B was highly responsive to salt stress. Morphological and physiological responses of rd29a and rd29b plants to salt stress were further investigated. Root growth, and photosynthetic properties declined significantly, while solute concentration (I pi), water use efficiency (WUE) and delta(13)C ratio increased under salt stress. Unexpectedly, the rd29a and rd29b knockout mutant lines maintained greater root growth, photosynthesis, and WUE under salt stress relative to control. We conclude that the RD29A and RD29B proteins are unlikely to serve directly as protective molecules.
DOI:10.1242/jcs.171207URLPMID:26395398 [本文引用: 1]

Seedlings of large-seeded plants are considered to be able to withstand abiotic stresses efficiently. The molecular mechanisms that underlie the involved signaling crosstalk between the large-seeded trait and abiotic tolerance are, however, largely unknown. Here, we demonstrate the molecular link that integrates plant abscisic acid (ABA) responses to drought stress into the regulation of seed mass. Both loss-of-function mutants of the Auxin Response Factor 2 (ARF2 encoding a transcription factor) and lines overexpressing AINTEGUMENTA (ANT; a transcription factor) under the 35S promoter exhibited large seed and drought-tolerant phenotypes as a result of abnormal ABA-auxin crosstalk signaling pathways in Arabidopsis. The target gene COLD-REGULATED15A (COR15a) was identified as participating in the regulation of seed development with ABA signaling through a negative regulation mechanism that is mediated by ANT. The molecular and genetic evidence presented indicate that ARF2, ANT and COR15A form an ABA-mediated signaling pathway to link modulation of seed mass with drought tolerance. These observations indicate that the ARF2 transcription factor serves as a molecular link that integrates plant ABA responses to drought stress into the regulation of seed mass.
