 ,, 郝玉金
,, 郝玉金 ,山东农业大学园艺科学与工程学院/作物生物学国家重点实验室/农业部黄淮地区园艺作物生物学与种质创制重点实验室,山东泰安 271018
,山东农业大学园艺科学与工程学院/作物生物学国家重点实验室/农业部黄淮地区园艺作物生物学与种质创制重点实验室,山东泰安 271018Analysis of Apple Ethylene Response Factor MdERF72 to Abiotic Stresses
WANG JiaHui, GU KaiDi, WANG ChuKun, YOU ChunXiang, HU DaGang ,, HAO YuJin
,, HAO YuJin ,College of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology/MOA Key Laboratory of Horticultural Crop Biology(Huanghuai Region)and Germplasm Innovation, Tai’an 271018, Shandong
,College of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology/MOA Key Laboratory of Horticultural Crop Biology(Huanghuai Region)and Germplasm Innovation, Tai’an 271018, Shandong通讯作者:
责任编辑: 赵伶俐
收稿日期:2019-03-21接受日期:2019-07-1网络出版日期:2019-12-01
| 基金资助: |
Received:2019-03-21Accepted:2019-07-1Online:2019-12-01
作者简介 About authors
王佳慧,E-mail:wangjhedu@126.com

摘要
关键词:
Abstract
Keywords:
PDF (4807KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王佳慧, 顾凯迪, 王楚堃, 由春香, 胡大刚, 郝玉金. 苹果乙烯响应因子MdERF72对非生物胁迫的响应[J]. 中国农业科学, 2019, 52(23): 4374-4385 doi:10.3864/j.issn.0578-1752.2019.23.017
WANG JiaHui, GU KaiDi, WANG ChuKun, YOU ChunXiang, HU DaGang, HAO YuJin.
0 引言
【研究意义】植物在生长发育过程中会遭受许多环境胁迫,如土壤盐渍化导致的盐胁迫和极端天气引发的温度胁迫等。盐胁迫是自然界普遍存在的非生物胁迫之一,盐碱化土壤中NaCl和MgCl2等可溶性盐的过量积累对植物造成离子胁迫,从而影响植物的正常生长发育过程[1]。低温胁迫是植物栽培中常常遇到的一种胁迫,它对植物的影响主要体现在酶活性、膜系统、细胞失水等,轻则导致作物产量下降,严重时会导致植株死亡。前人研究发现,ERF类转录因子具有调控高盐、低温等胁迫相关基因表达的功能[2]。因此,研究ERF类转录因子对改良植物对盐及温度胁迫的耐性,从而提高作物的抗逆性具有重要的理论意义。【前人研究进展】ERF转录因子是AP2/ERF家族中的一个亚家族,每个成员都含有1个由大约60个氨基酸组成的非常保守的DNA结合域。ERF转录因子在多种植物中广泛存在,拟南芥中发现124个ERF家族基因[3],水稻中有139个ERF家族基因[4]。近年来,在烟草[5]、玉米[6]、番茄[7]等植物中也分离出这类家族基因。关于ERF转录因子调控机理及抗逆胁迫功能的研究,在模式植物拟南芥、水稻、大豆等植物中已经有大量报道。研究证明ERFs家族基因受高盐、机械损伤、低氧、温度、氧化胁迫等非生物胁迫诱导。许多ERF转录因子在不同植物中异源表达能增强植株对非生物胁迫的抗性[8]。其中,DREB蛋白对非生物胁迫响应的研究较为透彻。过表达拟南芥中的AtDREB1/CBF提高了油菜的抗寒性[9],相反,拟南芥CBF1或CBF3的RNAi株系削弱了植物的耐寒力[10]。在番茄、烟草和水稻中异源表达拟南芥基因AtDREB1/ CBF可以提高植物耐寒性[11,12,13,14]。玫瑰DREB1/CBFs亚家族基因在拟南芥中被冷胁迫迅速诱导[15]。蒺藜苜蓿DREB1C转入蒺藜状苜蓿和中国玫瑰中也分别提高了转基因植株对寒冷的耐受性[16]。在番茄中异位超表达烟草TERF2可以提高对低温胁迫的耐受能力,而反义表达TERF2的番茄植株则对低温耐受力降低[17]。在盐胁迫方面,植物ERF家族基因中甘蔗SodERF3[18]、水稻OsEREBP1和OsEREBP2[19]、大麦HvRAF[20]、甘薯IbERF1和IbERF2[21]等对盐胁迫均有响应。刘文奇等[22]研究发现,ERF类基因OPBP1转入烟草中可以提高抗盐能力。【本研究切入点】目前,虽然在拟南芥及水稻等植物中鉴定了ERF家族部分成员的功能,例如在抵抗辣椒疫霉病、黄瓜花叶病[23]等生物胁迫和高盐、低温、干旱等非生物胁迫方面发挥着重要功能,但是,苹果中有关ERF的相关报道较少。前期研究发现,小金海棠MxERF72负调控苹果缺铁响应[24],但苹果中MdERF72对非生物胁迫的响应尚未有研究。【拟解决的关键问题】本研究通过转基因技术获得MdERF72过表达苹果愈伤组织,在对MdERF72的表达量与高盐、低温等非生物胁迫之间的关联分析基础上,探究MdERF72对非生物胁迫的响应,进一步了解MdERF72的功能,为苹果砧木的遗传改良提供参考。1 材料与方法
试验于2018年在山东农业大学作物生物学国家重点实验室进行。1.1 植物材料与处理
2018年5月开始对树龄9年的实生‘嘎拉’的生长根、茎、叶片、花(初花期)和果实(花后60、90、120、150、180、210和230 d)进行取样。根主要取自当年生新生根,茎和叶取自于当年生春梢新梢及梢部叶片,花为初花期整个花序,为保证一致性,取样时所有材料均取自一棵树,并且3个不同样品作为重复。‘嘎拉’苹果组培苗培养在固体培养基[30 g·L-1蔗糖+4.7 g·L-1 MS+0.2 mg·L-1 indole-3-acetic acid(IAA)+0.5 mg·L-1 6-Benzylaminopurine(6-BA)+8 g·L-1琼脂(pH 6.0)]上。用50 μmol·L-1乙烯合成前体1-氨基环丙烷羧酸(1-aminocyclopropanecarboxylic acid,ACC)、100 μmol·L-1 NaCl和4℃低温分别处理生长状况良好的组培苗,分别于处理0、1、3、6、12和24 h取样,提取RNA并反转录后利用实时荧光定量PCR的方法检测MdERF72的表达水平。
‘王林’苹果愈伤组织培养在固体培养基[30 g·L-1蔗糖+4.74 g·L-1 MS+0.5 mg·L-1 6-BA+1 mg·L-1 2,4-D+8 g·L-1琼脂(pH 6.0)]上,黑暗培养,温度25℃。
1.2 总RNA的提取和定量分析
采用韩朋良等[25]的方法提取RNA并反转录得到cDNA。然后以cDNA为模板,以5′-CCACCATCCTC TTCACCG-3′和5′-CACCCGCCACCTCTTTCT-3′为特异性引物,检测苹果中MdERF72的表达水平。采用张全艳等[26]方法进行荧光定量PCR反应,每样本进行3次重复。最后采用2-??CT法进行定量数据分析。
1.3 MdERF72的克隆
以MdERF72-F(5′-ATGTGTGGTGGTGCTATCA TTTCCG-3′)和MdERF72-R(5′-ATACAGAAGCT GCCCTTGTTGCTG-3′)分别为上、下游引物,‘嘎拉’组培苗叶片的cDNA为模板进行PCR扩增。反应体系为25 μL:其中ddH2O 17.25 μL,dNTPs(2 mmol·L-1)2 μL,HiFi buffer I 2.5 μL,上、下游引物(10 μmol·L-1)各1 μL,cDNA模板1 μL,HiFiTaq DNA Polymerase 0.25 μL。1.4 MdERF72的生物信息学分析
利用NCBI数据库(1.5 植物表达载体构建及农杆菌介导的苹果愈伤组织遗传转化
参照HU等 [27]的方法对‘王林’苹果的愈伤组织进行遗传转化。1.6 NaCl和低温处理苹果愈伤组织相关指标测定
将生长良好的野生型和转基因‘王林’苹果愈伤组织在25℃常温及4℃低温下处理。在暗处培养,分别于0、5、10和15 d测量鲜重、丙二醛(MDA)含量、相对电导率、过氧化氢(H2O2)含量及超氧阴离子(OFR)含量。将‘王林’苹果野生型和过量表达愈伤组织分别铺于愈伤固体培养基、添加100 mmol·L-1 NaCl的愈伤固体培养基和添加150 mmol·L-1 NaCl的愈伤固体培养基,在暗处培养一段时间后,观察表型及检测相关指标。
苹果愈伤组织中MDA含量采用硫代巴比妥酸法[28]测定,相对电导率采用崔之益等[29]的方法。过氧化氢(H2O2)含量及超氧阴离子含量(OFR)分别用过氧化氢(H2O2)测试盒(科铭生物,苏州,中国)、超氧阴离子(OFR)试剂盒(科铭生物,苏州,中国)进行检测。
1.7 数据统计与分析
使用DPS统计软件Tukey法对试验数据进行差异显著性分析(P<0.05)。2 结果
2.1 MdERF72全长的克隆
以‘嘎拉’苹果组培苗cDNA为模板,MdERF72- F和MdERF72-R为上、下游引物进行基因克隆。结果(图1)显示,PCR扩增获得了1条约750 bp的条带。测序发现,该基因包含长为759 bp完整的开放阅读框。与预测结果一致,可用于后续试验。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1MdERF72 PCR扩增
Fig. 1PCR amplification of MdERF72
2.2 MdERF72生物信息学分析
在AppleGFDB(图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2MdERF72及其同源基因的氨基酸序列比对分析
Fig. 2Sequence alignment of MdERF72 in apple and its homologous
用MEGA5.0软件最大似然法,对NCBI登录的拟南芥AtERF-B2亚家族的5个蛋白与MDP0000128979进行系统进化分析。结果发现,苹果ERF蛋白(MDP0000128979)与拟南芥AtERF72(At3g16770)亲缘关系最近(图3),因此将该基因命名为MdERF72。
图3
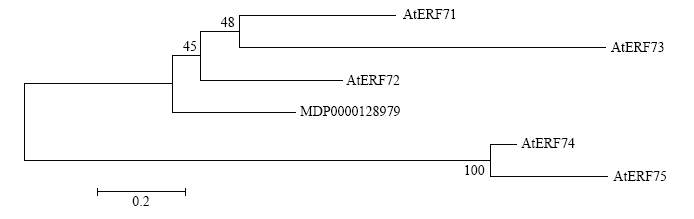 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3苹果ERF蛋白MDP0000128979与拟南芥ERF-B2家族蛋白系统进化树分析
Fig. 3Phylogenetic tree between apple ERF protein MDP0000756341 and Arabidopsis ERF-B2 family proteins
氨基酸理化性质分析表明,MdERF72编码253个氨基酸,预测其蛋白质分子量为27.61 kD,等电点(pI)为5.10。另外,亲疏水预测结果显示MdERF72疏水部分大于亲水部分,表明其属于疏水性蛋白。磷酸化位点分析显示,MdERF72只有苏氨酸磷酸化位点,表明该蛋白可能受到磷酸化作用的调控。
2.3 MdERF72启动子顺式作用元件分析
PlantCARE分析发现MdERF72启动子序列含有多个与抗逆性相关的调控元件:低温响应元件LTR,厌氧胁迫相关顺式作用元件ARE,干旱胁迫相关顺式作用元件MBS;另外,还发现了不同激素响应作用元件:生长素调节相关顺式作用元件TGA-box,茉莉酸响应顺式作用元件TGACG-motif core以及调节植物生长发育相关的光响应顺式作用元件Sp1(表1)。Table 1
表1
表1MdERF72基因启动子重要顺式作用元件分析
Table 1
| 调控序列 Regulatory sequence | 序列 Sequence | 位点功能 Function of site | 位置 Location |
|---|---|---|---|
| Sp1 | GGGCGG | 光响应元件Light responsive element | +51 |
| TGA-box | TGACGTAA | 生长素响应元件Part of an auxin-responsive element | -960 |
| LTR | CCGAAA | 低温响应元件cis-acting element involved in low-temperature responsiveness | -600 |
| ARE | AAACCA | 厌氧响应元件cis-acting regulatory element essential for the anaerobic induction | +1238 |
| MBS | CAACTG | 干旱胁迫响应元件MYB binding site involved in drought-inducibility | +1192 |
| TGACG-motif | TGACG | 茉莉酸响应元件cis-acting regulatory element involved in the MeJA-responsiveness | +768, -841 |
| CAT-box | GCCACT | 分生组织表达响应元件cis-acting regulatory element related to meristem expression | -1986 |
新窗口打开|下载CSV
2.4 苹果中MdERF72的表达分析
用50 μmol·L-1 ACC处理‘嘎拉’苹果组培苗后,实时荧光定量PCR检测MdERF72的相对表达水平。结果表明,MdERF72表达量相比对照明显升高,说明MdERF72是乙烯正调控转录因子(图4)。图4
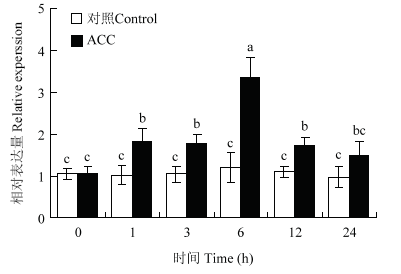 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4实时定量PCR检测‘嘎拉’组培苗中MdERF72对ACC的响应
不同字母在0.05水平上有显著差异。下同
Fig. 4Expression analysis of MdERF72 of Gala apple tissue culture seedlings in response to ACC
Different letters indicate significant difference at 0.05 levels. The same as below
通过实时定量PCR检测MdERF72在‘嘎拉’苹果生长根、茎、叶、花、果实(花后60 d)中的相对表达水平。结果表明,MdERF72在根、茎、叶、花、果实中均有表达,但茎和果实中的表达量较高(图5)。因此,检测了处于不同成熟阶段的果实中MdERF72的相对表达量。结果发现,随着果实成熟,MdERF72的表达量逐渐升高,在230 d达到顶峰(图6)。
图5
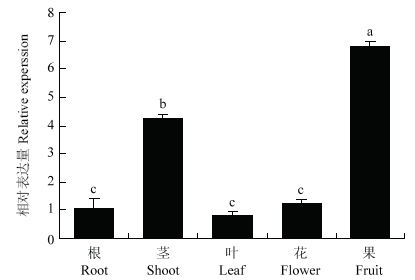 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5MdERF72在苹果不同组织中的相对表达水平
Fig. 5Expression of MdERF72 gene in different tissues of apple
图6
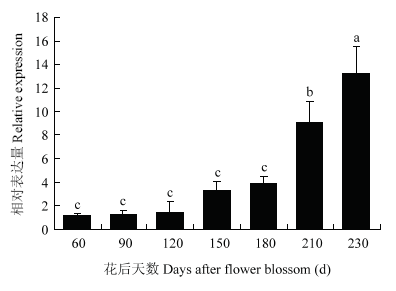 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6MdERF72在苹果不同发育时期的相对表达水平
Fig. 6Expression of MdERF72 gene in different developmental stages of apple
2.5 ‘嘎拉’组培苗中MdERF72对NaCl及4℃低温胁迫的响应
100 mmol·L-1 NaCl以及4℃低温处理‘嘎拉’苹果组培苗后,于0、1、3、6、12和24 h取样后进行qRT-PCR分析。结果表明,在NaCl以及4℃低温处理后,MdERF72随处理时间增加,表达量逐渐升高,在NaCl处理1 h达到最高值(图7-A),4℃低温处理6 h时达到顶峰(图7-B)。图7
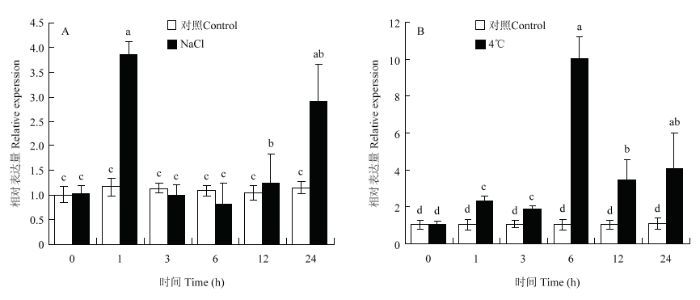 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7‘嘎拉’组培苗中MdERF72对NaCl(A)和4℃(B)低温的响应
Fig. 7Expression analysis of MdERF72 of Gala apple tissue culture seedlings in response to NaCl (A) and 4℃ (B) low temperature
2.6 农杆菌介导的苹果愈伤组织的遗传转化
构建35S::MdERF72过表达载体,通过农杆菌介导的方法侵染‘王林’苹果愈伤组织。获得3个转基因愈伤组织(图8),分别命名为MdERF72-OE1、MdERF72-OE2、MdERF72-OE3。图8
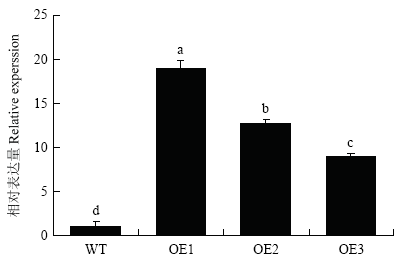 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8MdERF72转基因苹果愈伤组织的鉴定
Fig. 8Identification of MdERF72 transgenic apple calli
2.7 过量表达MdERF72苹果愈伤组织对NaCl及4℃低温胁迫的抗性
将MdERF72过量表达苹果愈伤组织(OE1、OE2、OE3)与野生型愈伤组织(WT)分别用0、100和150 mmol·L-1 NaCl的培养基处理。结果发现,在含有100和150 mmol·L-1 NaCl的培养基上培养15 d(图9),3个MdERF72过表达愈伤组织的生长速率均高于野生型(图10-A)、丙二醛含量(图10-B)、相对电导率(图10-C)、过氧化氢含量(图10-D)和超氧阴离子含量(图10-E)都低于野生型,表明MdERF72可以提高转基因苹果愈伤组织对高盐胁迫的抗性。图9
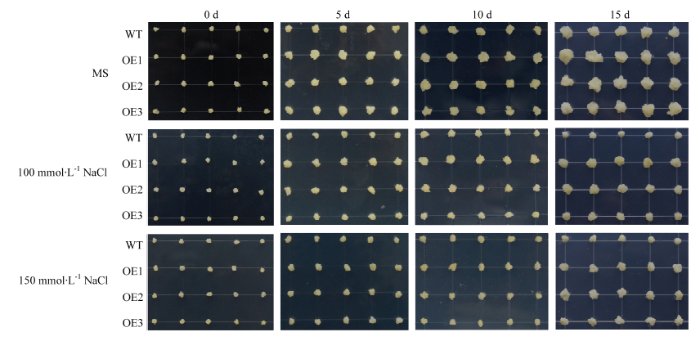 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9苹果野生型(WT)和MdERF72过量表达愈伤组织(OE1、OE2和OE3)在NaCl处理下的生长状态
Fig. 9Growth status of wild type (WT) and MdERF72 overexpressing apple calli (OE1, OE2 and OE3) under NaCl
图10
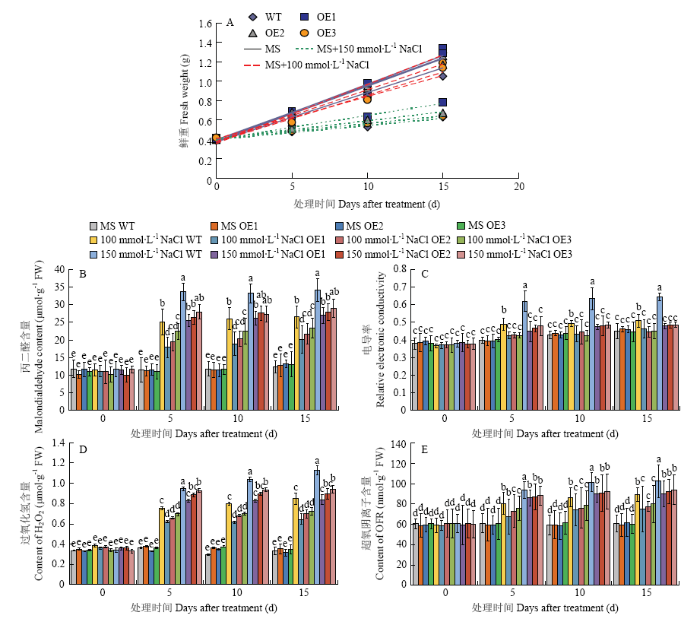 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10苹果野生型(WT)和MdERF72过量表达愈伤组织(OE1、OE2和OE3)在NaCl处理下的鲜重(A),丙二醛含量(B),电导率(C),过氧化氢含量(D),超氧阴离子含量(E)
Fig. 10Fresh weight (A), Malondialdehyde content (B), electronic conductivity (C), Hydrogen peroxide content(D) and Superoxide anion content (E) of wild type (WT) and MdERF72 overexpressing apple calli (OE1, OE2 and OE3) under NaCl treatments
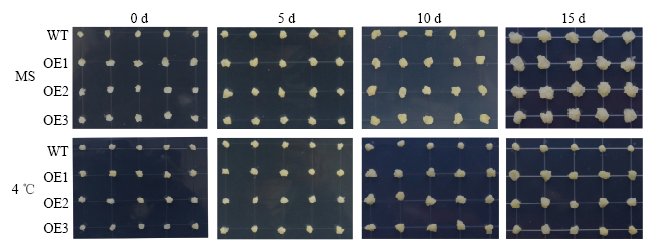
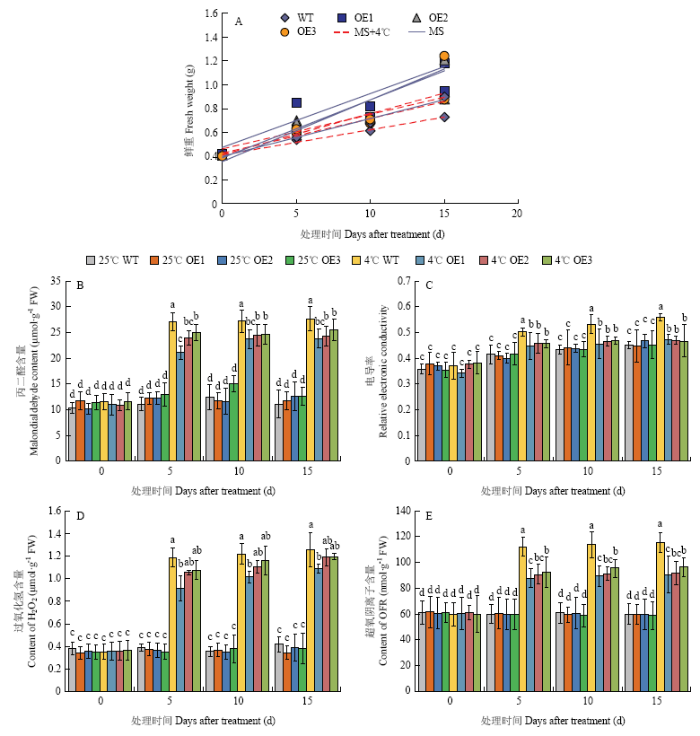
同样将MdERF72过量表达苹果愈伤组织(OE1、OE2和OE3)与野生型愈伤组织(WT)分别在25℃和4℃培养,结果发现,在4℃培养15 d后(图11),3个MdERF72过表达愈伤组织的生长速率都高于野生型(图12-A),丙二醛含量(图12-B)、相对电导率(图12-C)、过氧化氢含量(图12-D)和超氧阴离子含量(图12-E)都低于野生型,表明MdERF72转基因苹果愈伤组织对低温胁迫抗性提高。
图11
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图11苹果野生型(WT)和MdERF72过量表达愈伤组织(OE1、OE2和OE3)在4℃低温处理下的生长状态
Fig. 11Growth status of wild type (WT) and MdERF72 overexpressing apple calli (OE1, OE2 and OE3) under 4℃ low temperature
图12
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图12苹果野生型(WT)和MdERF72过量表达愈伤组织(OE1、OE2和OE3)在4℃低温处理下的鲜重(A),丙二醛含量(B),电导率(C),过氧化氢含量(D),超氧阴离子含量(E)
Fig. 12Fresh weight (A), Malondialdehyde content (B), electronic conductivity (C), Hydrogen peroxide content (D) and Superoxide anion content (E) of wild type (WT) and MdERF72 overexpressing apple calli (OE1, OE2 and OE3) under 4℃ low temperature
3 讨论
苹果是世界上最重要的经济作物之一。中国是世界上苹果种植面积最大、产量最高的国家,随着国际农业合作力度的不断加深,我国也成为世界上苹果及相关加工品主要出口国之一。然而,近年来极端天气频发,苹果的生长面临着严峻的考验,其中盐碱化、干旱、低温等胁迫对苹果的产量和品质造成了严重损伤,因此,选育抗盐、抗旱、耐寒的优良苹果砧木是目前所要解决的关键问题。面对复杂的环境变化,植物通过一系列的信号传递和能量代谢抵御外界不良环境的侵害。在这一过程中,转录因子发挥着重要的调控作用。因此,研究转录因子的功能,对提高植物的抗逆性及选育优良品种具有重要意义。乙烯应答因子是植物特有的一类转录因子,与生物/非生物胁迫密切相关。例如,在水稻和菊花中过表达AtDREB1A,可以提高转基因植株在干旱、高盐和低温胁迫处理下的耐受性[30,31,32];在烟草中过表达NtERF5可以提高对烟草花叶病毒的抗性[33];在烟草中过表达陆地棉GbERF2可以提高对赤星病的抗性[34]。关于苹果中ERF的功能,前人研究发现,苹果MdERF2参与茉莉酸(JA)介导的果实成熟[35]。然而,苹果中ERFs对非生物胁迫的响应尚未有报道。本研究发现,MdERF72的表达受高盐和低温胁迫诱导;并且随着果实的成熟,内源乙烯释放量增多,MdERF72的表达量逐渐升高,因此,推断MdERF72是乙烯响应的正调控因子。另外,本研究表明,MdERF72除在苹果茎中表达量较高外,在果实中的表达量也很高,可能参与苹果果实抵御外界不利的非生
物和生物胁迫;而且前人的研究表明,乙烯响应因子作为激素和胁迫信号的一个非常重要的调节中心[36],这一报道支持乙烯响应因子家族基因MdERF72可能参与非生物胁迫抵抗力;同时,本研究发现在苹果愈伤组织中过表达MdERF72确实可以提高其对高盐和低温胁迫的抗性。
提高作物对盐胁迫的抗性,可以有效的提高产量和品质。由于气候多变、植被破坏加剧及耕作管理不当等原因,土壤盐碱化越来越严重,据不完全统计,全球9亿多hm2土地面临土壤盐碱化问题。土壤盐碱化会导致植物根系对水分和营养的吸收受到抑制,使树势衰弱,抗性下降,病害频发。面对土壤盐碱化,除了从改良土壤方面入手,还可以通过提高植物的耐盐性来改善。在水稻中,植物对盐胁迫的响应机制已经有较为完善的研究。在苹果中,植物对盐胁迫的响应尚不明确。本研究中发现的MdERF72正调控苹果对高盐及低温的胁迫响应,对于选育抗盐、耐寒的优良苹果砧木具有重要指导意义。
4 结论
克隆获得苹果乙烯响应因子基因MdERF72,该基因编码253个氨基酸。MdERF72包含一个AP2保守结构域。进化树分析显示,MdERF72与拟南芥AtERF72亲缘关系最近。MdERF72在苹果茎和果实中表达量较高,并且对高盐和低温胁迫有明显响应。MdERF72转基因愈伤在高盐和低温胁迫下生长势要明显强于野生型,说明MdERF72在响应高盐和低温胁迫过程中发挥着重要的正调控作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[D].
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/nph.12291URLPMID:24010138 [本文引用: 1]

Transcription factors belonging to the APETALA2/Ethylene Responsive Factor (AP2/ERF) family are conservatively widespread in the plant kingdom. These regulatory proteins are involved in the control of primary and secondary metabolism, growth and developmental programs, as well as responses to environmental stimuli. Due to their plasticity and to the specificity of individual members of this family, AP2/ERF transcription factors represent valuable targets for genetic engineering and breeding of crops. In this review, we integrate the evidence collected from functional and structural studies to describe their different mechanisms of action and the regulatory pathways that affect their activity.
DOI:10.1126/science.290.5499.2105URLPMID:11118137 [本文引用: 1]

The completion of the Arabidopsis thaliana genome sequence allows a comparative analysis of transcriptional regulators across the three eukaryotic kingdoms. Arabidopsis dedicates over 5% of its genome to code for more than 1500 transcription factors, about 45% of which are from families specific to plants. Arabidopsis transcription factors that belong to families common to all eukaryotes do not share significant similarity with those of the other kingdoms beyond the conserved DNA binding domains, many of which have been arranged in combinations specific to each lineage. The genome-wide comparison reveals the evolutionary generation of diversity in the regulation of transcription.
DOI:10.1104/pp.105.073783URLPMID:16407444 [本文引用: 1]

Genes in the ERF family encode transcriptional regulators with a variety of functions involved in the developmental and physiological processes in plants. In this study, a comprehensive computational analysis identified 122 and 139 ERF family genes in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa L. subsp. japonica), respectively. A complete overview of this gene family in Arabidopsis is presented, including the gene structures, phylogeny, chromosome locations, and conserved motifs. In addition, a comparative analysis between these genes in Arabidopsis and rice was performed. As a result of these analyses, the ERF families in Arabidopsis and rice were divided into 12 and 15 groups, respectively, and several of these groups were further divided into subgroups. Based on the observation that 11 of these groups were present in both Arabidopsis and rice, it was concluded that the major functional diversification within the ERF family predated the monocot/dicot divergence. In contrast, some groups/subgroups are species specific. We discuss the relationship between the structure and function of the ERF family proteins based on these results and published information. It was further concluded that the expansion of the ERF family in plants might have been due to chromosomal/segmental duplication and tandem duplication, as well as more ancient transposition and homing. These results will be useful for future functional analyses of the ERF family genes.
DOI:10.1105/tpc.7.2.173URLPMID:7756828 [本文引用: 1]

We demonstrated that the GCC box, which is an 11-bp sequence (TAAGAGCCGCC) conserved in the 5' upstream region of ethylene-inducible pathogenesis-related protein genes in Nicotiana spp and in some other plants, is the sequence that is essential for ethylene responsiveness when incorporated into a heterologous promoter. Competitive gel retardation assays showed DNA binding activities to be specific to the GCC box sequence in tobacco nuclear extracts. Four different cDNAs encoding DNA binding proteins specific for the GCC box sequence were isolated, and their products were designated ethylene-responsive element binding proteins (EREBPs). The deduced amino acid sequences of EREBPs exhibited no homology with those of known DNA binding proteins or transcription factors; neither did the deduced proteins contain a basic leucine zipper or zinc finger motif. The DNA binding domain was identified within a region of 59 amino acid residues that was common to all four deduced EREBPs. Regions highly homologous to the DNA binding domain of EREBPs were found in proteins deduced from the cDNAs of various plants, suggesting that this domain is evolutionarily conserved in plants. RNA gel blot analysis revealed that accumulation of mRNAs for EREBPs was induced by ethylene, but individual EREBPs exhibited different patterns of expression.
DOI:10.1055/s-0039-3400233URLPMID:31842235 [本文引用: 1]

Thoracic aortic aneurysm is a typically silent disease characterized by a lethal natural history. Since the discovery of the familial nature of thoracic aortic aneurysm and dissection (TAAD) almost 2 decades ago, our understanding of the genetics of this disorder has undergone a transformative amplification. To date, at least 37 TAAD-causing genes have been identified and an estimated 30% of the patients with familial nonsyndromic TAAD harbor a pathogenic mutation in one of these genes. In this review, we present our yearly update summarizing the genes associated with TAAD and the ensuing clinical implications for surgical intervention. Molecular genetics will continue to bolster this burgeoning catalog of culprit genes, enabling the provision of personalized aortic care.
DOI:10.1105/tpc.000794URLPMID:11971137 [本文引用: 1]

The Pti4, Pti5, and Pti6 proteins from tomato were identified based on their interaction with the product of the Pto disease resistance gene, a Ser-Thr protein kinase. They belong to the ethylene-response factor (ERF) family of plant-unique transcription factors and bind specifically to the GCC-box cis element present in the promoters of many pathogenesis-related (PR) genes. Here, we show that these tomato ERFs are localized to the nucleus and function in vivo as transcription activators that regulate the expression of GCC box-containing PR genes. Expression of Pti4, Pti5, or Pti6 in Arabidopsis activated the expression of the salicylic acid-regulated genes PR1 and PR2. Expression of jasmonic acid- and ethylene-regulated genes, such as PR3, PR4, PDF1.2, and Thi2.1, was affected differently by each of the three tomato ERFs, with Arabidopsis-Pti4 plants having very high levels of PDF1.2 transcripts. Exogenous application of salicylic acid to Arabidopsis-Pti4 plants suppressed the increased expression of PDF1.2 but further stimulated PR1 expression. Arabidopsis plants expressing Pti4 displayed increased resistance to the fungal pathogen Erysiphe orontii and increased tolerance to the bacterial pathogen Pseudomonas syringae pv tomato. These results indicate that Pti4, Pti5, and Pti6 activate the expression of a wide array of PR genes and play important and distinct roles in plant defense.
DOI:10.1111/j.1467-7652.2010.00578.xURL [本文引用: 1]

We show here that transgenic Arabidopsis plants that expressed chimeric repressors derived from the AtMYB102, ANAC047, HRS1, ZAT6 and AtERF5 transcription factors were tolerant to treatment with 400 mM NaCl, which was lethal to wild-type plants. The transgenic plants grew well, without any apparent differences from the wild-type plants under normal growth condition. The transgenic lines expressing the AtMYB102, ANAC047 and HRS1 chimeric repressors germinated in the presence of 225 mM NaCl, while those expressing the ZAT6 and AtERF5 did not. However, the latter lines were tolerant to osmotic stress and germinated in the presence of 600 mM mannitol, suggesting a link between responses to salt and osmotic stress. Expression of the AtMYB102, ANAC047, ZAT6 and AtERF5 genes was induced by salt treatment, while that of HRS1 was repressed. HRS1 has transcriptional repressive activity and appears to suppress the expression of factors that negatively regulate salt tolerance. Microarray analysis revealed that the levels of expression of DREB1A, DREB2B and several genes for ZAT transcription factors rose 10-to 100-fold in the AtMYB102 chimeric repressor line under both normal and stress conditions. Elevated expression of DREB- and ZAT-related genes might be involved in the salt tolerance of the AtMYB102 chimeric repressor line. Transgenic rice plants expressing chimeric repressors derived from Os02g0325600 and Os03g0327800, rice homologues of HRS1 and ANAC047, were tolerant to salinity stress demonstrated by suppression of growth inhibition and ion leakages. Expression of a chimeric repressor provides an effective strategy for enhancing tolerance of plants to abiotic stress.
URLPMID:11706173 [本文引用: 1]

Many plants increase in freezing tolerance in response to low, nonfreezing temperatures, a phenomenon known as cold acclimation. Cold acclimation in Arabidopsis involves rapid cold-induced expression of the C-repeat/dehydration-responsive element binding factor (CBF) transcriptional activators followed by expression of CBF-targeted genes that increase freezing tolerance. Here, we present evidence for a CBF cold-response pathway in Brassica napus. We show that B. napus encodes CBF-like genes and that transcripts for these genes accumulate rapidly in response to low temperature followed closely by expression of the cold-regulated Bn115 gene, an ortholog of the Arabidopsis CBF-targeted COR15a gene. Moreover, we show that constitutive overexpression of the Arabidopsis CBF genes in transgenic B. napus plants induces expression of orthologs of Arabidopsis CBF-targeted genes and increases the freezing tolerance of both nonacclimated and cold-acclimated plants. Transcripts encoding CBF-like proteins were also found to accumulate rapidly in response to low temperature in wheat (Triticum aestivum L. cv Norstar) and rye (Secale cereale L. cv Puma), which cold acclimate, as well as in tomato (Lycopersicon esculentum var. Bonny Best, Castle Mart, Micro-Tom, and D Huang), a freezing-sensitive plant that does not cold acclimate. An alignment of the CBF proteins from Arabidopsis, B. napus, wheat, rye, and tomato revealed the presence of conserved amino acid sequences, PKK/RPAGRxKFxETRHP and DSAWR, that bracket the AP2/EREBP DNA binding domains of the proteins and distinguish them from other members of the AP2/EREBP protein family. We conclude that components of the CBF cold-response pathway are highly conserved in flowering plants and not limited to those that cold acclimate.
DOI:10.1073/pnas.0705639105URLPMID:18093929 [本文引用: 1]

The C-repeat-binding factor (CBF)/dehydration-responsive element-binding factor (DREB1) proteins constitute a small family of Arabidopsis transcriptional activators (CBF1/DREB1B, CBF2/DREB1C, and CBF3/DREB1A) that play a prominent role in cold acclimation. A fundamental question about these factors that remains to be answered is whether they are functionally equivalent. Recently, we reported that CBF2 negatively regulates CBF1 and CBF3 expression, and that CBFs are subjected to different temporal regulation during cold acclimation, which suggested this might not be the case. In this study, we have analyzed the expression of CBF genes in different tissues of Arabidopsis, during development and in response to low temperature, and characterized RNA interference (RNAi) and antisense lines that fail to accumulate CBF1 or/and CBF3 mRNAs under cold conditions. We found that CBF1 and CBF3 are regulated in a different way than CBF2. Moreover, in contrast to CBF2, CBF1 and CBF3 are not involved in regulating other CBF genes and positively regulate cold acclimation by activating the same subset of CBF-target genes. All these results demonstrate that CBF1 and CBF3 have different functions than CBF2. We also found that the CBF regulon is composed of at least two different kind of genes, one of them requiring the simultaneous expression of both CBF1 and CBF3 to be properly induced. This indicates that CBF1 and CBF3 have a concerted additive effect to induce the whole CBF regulon and the complete development of cold acclimation.
DOI:10.1104/pp.003442URLPMID:12114563 [本文引用: 1]

In an attempt to improve stress tolerance of tomato (Lycopersicon esculentum) plants, an expression vector containing an Arabidopsis C-repeat/dehydration responsive element binding factor 1 (CBF1) cDNA driven by a cauliflower mosaic virus 35S promoter was transferred into tomato plants. Transgenic expression of CBF1 was proved by northern- and western-blot analyses. The degree of chilling tolerance of transgenic T(1) and T(2) plants was found to be significantly greater than that of wild-type tomato plants as measured by survival rate, chlorophyll fluorescence value, and radical elongation. The transgenic tomato plants exhibited patterns of growth retardation; however, they resumed normal growth after GA(3) (gibberellic acid) treatment. More importantly, GA(3)-treated transgenic plants still exhibited a greater degree of chilling tolerance compared with wild-type plants. Subtractive hybridization was performed to isolate the responsive genes of heterologous Arabidopsis CBF1 in transgenic tomato plants. CATALASE1 (CAT1) was obtained and showed activation in transgenic tomato plants. The CAT1 gene and catalase activity were also highly induced in the transgenic tomato plants. The level of H(2)O(2) in the transgenic plants was lower than that in the wild-type plants under either normal or cold conditions. The transgenic plants also exhibited considerable tolerance against oxidative damage induced by methyl viologen. Results from the current study suggest that heterologous CBF1 expression in transgenic tomato plants may induce several oxidative-stress responsive genes to protect from chilling stress.
DOI:10.1093/pcp/pch037URLPMID:15047884 [本文引用: 1]

The transcription factor DREB1A/CBF3 specifically interacts with the dehydration responsive element (DRE/CRT) and induces expression of genes involved in environmental stress tolerance in Arabidopsis: Overexpression of DREB1A improved drought- and low-temperature stress tolerance in tobacco. The stress-inducible rd29A promoter minimized the negative effects on the plant growth in tobacco. Furthermore, we detected overexpression of stress-inducible target genes of DREB1A in tobacco. These results indicate that a combination of the rd29A promoter and DREB1A is useful for improvement of various kinds of transgenic plants that are tolerant to environmental stress.
DOI:10.1093/pcp/pci230URLPMID:16284406 [本文引用: 1]

The transcription factors dehydration-responsive element-binding protein 1s (DREB1s)/C-repeat-binding factors (CBFs) specifically interact with the DRE/CRT cis-acting element and control the expression of many stress-inducible genes in Arabidopsis. The genes for DREB1 orthologs, OsDREB1A and OsDREB1B from rice, are induced by cold stress, and overexpression of DREB1 or OsDREB1 induced strong expression of stress-responsive genes in transgenic Arabidopsis plants, resulting in increased tolerance to high-salt and freezing stresses. In this study, we generated transgenic rice plants overexpressing the OsDREB1 or DREB1 genes. These transgenic rice plants showed not only growth retardation under normal growth conditions but also improved tolerance to drought, high-salt and low-temperature stresses like the transgenic Arabidopsis plants overexpressing OsDREB1 or DREB1. We also detected elevated contents of osmoprotectants such as free proline and various soluble sugars in the transgenic rice as in the transgenic Arabidopsis plants. We identified target stress-inducible genes of OsDREB1A in the transgenic rice using microarray and RNA gel blot analyses. These genes encode proteins that are thought to function in stress tolerance in the plants. These results indicate that the DREB1/CBF cold-responsive pathway is conserved in rice and the DREB1-type genes are quite useful for improvement of stress tolerance to environmental stresses in various kinds of transgenic plants including rice.
DOI:10.1111/j.1365-3040.2008.01776.xURLPMID:18182016 [本文引用: 1]

We studied the effect of ectopic AtCBF over-expression on physiological alterations that occur during cold exposure in frost-sensitive Solanum tuberosum and frost-tolerant Solanum commersonii. Relative to wild-type plants, ectopic AtCBF1 over-expression induced expression of COR genes without a cold stimulus in both species, and imparted a significant freezing tolerance gain in both species: 2 degrees C in S. tuberosum and up to 4 degrees C in S. commersonii. Transgenic S. commersonii displayed improved cold acclimation potential, whereas transgenic S. tuberosum was still incapable of cold acclimation. During cold treatment, leaves of wild-type S. commersonii showed significant thickening resulting from palisade cell lengthening and intercellular space enlargement, whereas those of S. tuberosum did not. Ectopic AtCBF1 activity induced these same leaf alterations in the absence of cold in both species. In transgenic S. commersonii, AtCBF1 activity also mimicked cold treatment by increasing proline and total sugar contents in the absence of cold. Relative to wild type, transgenic S. commersonii leaves were darker green, had higher chlorophyll and lower anthocyanin levels, greater stomatal numbers, and displayed greater photosynthetic capacity, suggesting higher productivity potential. These results suggest an endogenous CBFpathway is involved in many of the structural, biochemical and physiological alterations associated with cold acclimation in these Solanum species.
DOI:10.1105/tpc.10.8.1391URLPMID:9707537 [本文引用: 1]

Plant growth is greatly affected by drought and low temperature. Expression of a number of genes is induced by both drought and low temperature, although these stresses are quite different. Previous experiments have established that a cis-acting element named DRE (for dehydration-responsive element) plays an important role in both dehydration- and low-temperature-induced gene expression in Arabidopsis. Two cDNA clones that encode DRE binding proteins, DREB1A and DREB2A, were isolated by using the yeast one-hybrid screening technique. The two cDNA libraries were prepared from dehydrated and cold-treated rosette plants, respectively. The deduced amino acid sequences of DREB1A and DREB2A showed no significant sequence similarity, except in the conserved DNA binding domains found in the EREBP and APETALA2 proteins that function in ethylene-responsive expression and floral morphogenesis, respectively. Both the DREB1A and DREB2A proteins specifically bound to the DRE sequence in vitro and activated the transcription of the b-glucuronidase reporter gene driven by the DRE sequence in Arabidopsis leaf protoplasts. Expression of the DREB1A gene and its two homologs was induced by low-temperature stress, whereas expression of the DREB2A gene and its single homolog was induced by dehydration. Overexpression of the DREB1A cDNA in transgenic Arabidopsis plants not only induced strong expression of the target genes under unstressed conditions but also caused dwarfed phenotypes in the transgenic plants. These transgenic plants also revealed freezing and dehydration tolerance. In contrast, overexpression of the DREB2A cDNA induced weak expression of the target genes under unstressed conditions and caused growth retardation of the transgenic plants. These results indicate that two independent families of DREB proteins, DREB1 and DREB2, function as trans-acting factors in two separate signal transduction pathways under low-temperature and dehydration conditions, respectively.
DOI:10.1007/s10725-009-9434-4URL [本文引用: 1]

We isolated a DREB orthologue, MtDREB1C, from Medicago truncatula. Its deduced protein contains an AP2 domain of 57 amino acids. Yeast one-hybrid assay revealed that MtDREB1C specifically bound to the dehydration-responsive element (DRE) and activated the expression of HIS3 and LacZ reporter genes. In a transcriptional activation assay, coexpression of the MtDREB1C cDNA resulted in much higher (21.2 times) transactivation of the LacZ reporter gene than experiments performed without MtDREB1C. Transformation of Medicago revealed that overexpression of MtDREB1C suppressed shoot growth, and enhanced the freezing tolerance of M. truncatula. The MtDREB1C gene was transformed into China Rose (Rosa chinensis Jacq.) driven by Arabidopsis rd29A promoter. Southern-blot analysis showed that the target gene was integrated into the genome of a surviving transgenic rose plant. Northern-blot analysis illustrated that robust expression of MtDREB1C was only activated under stress conditions, and the expressed MtDREB1C mRNA reached maximum accumulation 10h following freezing treatment. The performance of the transgenic line under freezing stress was superior to untransformed controls. This transgenic plant continued to grow, flowered under unstressed conditions, and was phenotypically normal. These facts indicate that the MtDREB1C gene, isolated from Medicago truncatula and driven by the Arabidopsis rd29A promoter, enhanced freezing tolerance in transgenic China Rose significantly without any obvious morphological or developmental abnormality.
DOI:10.1007/s11103-010-9609-4URL [本文引用: 1]

Increasing numbers of investigations indicate that ethylene response factor (ERF) proteins play important roles in plant stress responses via interacting with GCC box and/dehydration-responsive element/C-repeat to modulate expression of downstream genes, but the detailed regulatory mechanism is not well elucidated. Revealing the modulation pathway of ERF proteins in response to stresses is vital. Previously, we showed that tomato ERF protein TERF2/LeERF2 is ethylene inducible, and ethylene production is suppressed in antisense TERF2/LeERF2 tomatoes, suggesting that TERF2/LeERF2 functions as a positive regulator in ethylene biosynthesis. In this paper, we report that regulation of TERF2/LeERF2 in ethylene biosynthesis is associated with enhanced freezing tolerance of tobacco and tomato. Analysis of gene expression showed that cold slowly induces expression of TERF2/LeERF2 in tomato, implying that TERF2/LeERF2 may be involved in cold response through ethylene modulation. To test the hypothesis, we first observed that overexpressing TERF2/LeERF2 tobaccos not only enhances freezing tolerance via activating expression of cold-related genes, but also significantly reduces electrolyte leakage. In addition, with treatment of ethylene biosynthesis inhibitor or ethylene receptor antagonist, we then showed that blockage of ethylene biosynthesis or the ethylene signaling pathway decreases freezing tolerance of overexpressing TERF2/LeERF2 tobaccos. Moreover, the results from tomatoes showed that overexpressing TERF2/LeERF2 tomatoes enhances while antisense TERF2/LeERF2 transgenic lines decreases freezing tolerance, and application of ethylene precursor 1-aminocyclopropane-1-carboxylic acid restored freezing tolerance of antisense lines. Therefore our results establish that TERF2/LeERF2 enhances freezing tolerance of plants through ethylene biosynthesis and the ethylene signaling pathway.
DOI:10.1093/pcp/pcn025URLPMID:18281696 [本文引用: 1]

The molecular signals and pathways that govern biotic and abiotic stress responses in sugarcane are poorly understood. Here we describe SodERF3, a sugarcane (Saccharum officinarum L. cv Ja60-5) cDNA that encodes a 201-amino acid DNA-binding protein that acts as a transcriptional regulator of the ethylene responsive factor (ERF) superfamily. Like other ERF transcription factors, the SodERF3 protein binds to the GCC box, and its deduced amino acid sequence contains an N-terminal putative nuclear localization signal (NLS). In addition, a C-terminal short hydrophobic region that is highly homologous to an ERF-associated amphiphilic repression-like motif, typical for class II ERFs, was found. Northern and Western blot analysis showed that SodERF3 is induced by ethylene. In addition, SodERF3 is induced by ABA, salt stress and wounding. Greenhouse-grown transgenic tobacco plants (Nicotiana tabacum L. cv. SR1) expressing SodERF3 were found to display increased tolerance to drought and osmotic stress.
DOI:10.1007/s11103-013-0073-9URLPMID:23703395 [本文引用: 1]

High salinity causes remarkable losses in rice productivity worldwide mainly because it inhibits growth and reduces grain yield. To cope with environmental changes, plants evolved several adaptive mechanisms, which involve the regulation of many stress-responsive genes. Among these, we have chosen OsRMC to study its transcriptional regulation in rice seedlings subjected to high salinity. Its transcription was highly induced by salt treatment and showed a stress-dose-dependent pattern. OsRMC encodes a receptor-like kinase described as a negative regulator of salt stress responses in rice. To investigate how OsRMC is regulated in response to high salinity, a salt-induced rice cDNA expression library was constructed and subsequently screened using the yeast one-hybrid system and the OsRMC promoter as bait. Thereby, two transcription factors (TFs), OsEREBP1 and OsEREBP2, belonging to the AP2/ERF family were identified. Both TFs were shown to bind to the same GCC-like DNA motif in OsRMC promoter and to negatively regulate its gene expression. The identified TFs were characterized regarding their gene expression under different abiotic stress conditions. This study revealed that OsEREBP1 transcript level is not significantly affected by salt, ABA or severe cold (5 °C) and is only slightly regulated by drought and moderate cold. On the other hand, the OsEREBP2 transcript level increased after cold, ABA, drought and high salinity treatments, indicating that OsEREBP2 may play a central role mediating the response to different abiotic stresses. Gene expression analysis in rice varieties with contrasting salt tolerance further suggests that OsEREBP2 is involved in salt stress response in rice.
DOI:10.1007/s00425-006-0373-2URL [本文引用: 1]

We isolated HvRAF (Hordeum vulgare root abundant factor), a cDNA encoding a novel ethylene response factor (ERF)-type transcription factor, from young seedlings of barley. In addition to the most highly conserved APETALA2/ERF DNA-binding domain, the encoded protein contained an N-terminal MCGGAIL signature sequence, a putative nuclear localization sequence, and a C-terminal acidic transcription activation domain containing a novel mammalian hemopexin domain signature-like sequence. Their homologous sequences were found in AAK92635 from rice and RAP2.2 from Arabidopsis; the ERF proteins most closely related to HvRAF, reflecting their functional importance. RNA blot analyses revealed that HvRAF transcripts were more abundant in roots than in leaves. HvRAF expression was induced in barley seedlings by various treatment regimes such as salicylic acid, ethephon, methyl jasmonate, cellulase, and methyl viologen. In a subcellular localization assay, the HvRAF-GFP fusion protein was targeted to the nucleus. The fusion protein of HvRAF with the GAL4 DNA-binding domain strongly activated transcription in yeast. Various deletion mutants of HvRAF indicated that the transactivating activity was localized to the acidic domain of the C-terminal region, and that the hemopexin domain signature-like sequence was important for the activity. Overexpression of the HvRAF gene in Arabidopsis plants induced the activation of various stress-responsive genes, including PDF1.2, JR3, PR1, PR5, KIN2, and GSH1. Furthermore, the transgenic Arabidopsis plants showed enhanced resistance to Ralstonia solanacearum strain GMI1000, as well as seed germination and root growth tolerance to high salinity. These results collectively indicate that HvRAF is a transcription factor that plays dual regulatory roles in response to biotic and abiotic stresses in plants.
DOI:10.1016/j.jplph.2012.03.002URL [本文引用: 1]

Two ethylene response factor (ERF) genes, IbERF1 and IbERF2, were isolated from a library of expressed sequence tags (EST) prepared from suspension-cultured cells and dehydration-treated fibrous roots of sweetpotato (Ipomoea batatas). The deduced IbERFs contained a nuclear localization signal and the AP2/ERF DNA-binding domain. RT-PCR analysis revealed that IbERF1 was expressed abundantly during the growth of suspension-cultured cells, whereas the expression levels of IbERF2 transcripts were high in fibrous, thick pigmented roots. Two ERF genes also showed different responses to various types of abiotic stress and pathogen infection. Transient expression of the two ERF genes in tobacco (Nicotiana tabacum) leaves resulted in increased transcript levels of the pathogenesis-related 5 (PR5) gene, the early response to dehydration ten gene (ERD10), the CuZn superoxide dismutase gene (CuZnSOD) and the catalase gene (CAT). It is suggested that the two ERF genes play roles in the stress defense-signaling pathway as transcriptional regulators of the PR5, ERD10, CuZnSOD and CAT genes. (C) 2012 Elsevier GmbH.
[本文引用: 1]
[本文引用: 1]
DOI:10.1094/MPMI.2002.15.10.983URLPMID:12437295 [本文引用: 1]

In many plants, including hot pepper plants, productivity is greatly affected by pathogen attack. We reported previously that tobacco stress-induced gene 1 (Tsi1) may play an important role in regulating stress responsive genes and pathogenesis-related (PR) genes. In this study, we demonstrated that overexpression of Tsi1 gene in transgenic hot pepper plants induced constitutive expression of several PR genes in the absence of stress or pathogen treatment. The transgenic hot pepper plants expressing Tsi1 exhibited resistance to Pepper mild mottle virus (PMMV) and Cucumber mosaic virus (CMV). Furthermore, these transgenic plants showed increased resistance to a bacterial pathogen, Xanthomonas campestris pv. vesicatoria and also an oomycete pathogen, Phytophthora capsici. These results suggested that ectopic expression of Tsi1 in transgenic hot pepper plants enhanced the resistance of the plants to various pathogens, including viruses, bacteria, and oomycete. These results suggest that using transcriptional regulatory protein genes may contribute to developing broad-spectrum resistance in crop plants.
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.15.01333URLPMID:26637549 [本文引用: 1]

Tonoplast transporters, including proton pumps and secondary transporters, are essential for plant cell function and for quality formation of fleshy fruits and ornamentals. Vacuolar transport of anthocyanins, malate, and other metabolites is directly or indirectly dependent on the H(+)-pumping activities of vacuolar H(+)-ATPase (VHA) and/or vacuolar H(+)-pyrophosphatase, but how these proton pumps are regulated in modulating vacuolar transport is largely unknown. Here, we report a transcription factor, MdMYB1, in apples that binds to the promoters of two genes encoding the B subunits of VHA, MdVHA-B1 and MdVHA-B2, to transcriptionally activate its expression, thereby enhancing VHA activity. A series of transgenic analyses in apples demonstrates that MdMYB1/10 controls cell pH and anthocyanin accumulation partially by regulating MdVHA-B1 and MdVHA-B2. Furthermore, several other direct target genes of MdMYB10 are identified, including MdVHA-E2, MdVHP1, MdMATE-LIKE1, and MdtDT, which are involved in H(+)-pumping or in the transport of anthocyanins and malates into vacuoles. Finally, we show that the mechanism by which MYB controls malate and anthocyanin accumulation in apples also operates in Arabidopsis (Arabidopsis thaliana). These findings provide novel insights into how MYB transcription factors directly modulate the vacuolar transport system in addition to anthocyanin biosynthesis, consequently controlling organ coloration and cell pH in plants.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.104.059147URLPMID:15834008 [本文引用: 1]

Rice (Oryza sativa), a monocotyledonous plant that does not cold acclimate, has evolved differently from Arabidopsis (Arabidopsis thaliana), which cold acclimates. To understand the stress response of rice in comparison with that of Arabidopsis, we developed transgenic rice plants that constitutively expressed CBF3/DREB1A (CBF3) and ABF3, Arabidopsis genes that function in abscisic acid-independent and abscisic acid-dependent stress-response pathways, respectively. CBF3 in transgenic rice elevated tolerance to drought and high salinity, and produced relatively low levels of tolerance to low-temperature exposure. These data were in direct contrast to CBF3 in Arabidopsis, which is known to function primarily to enhance freezing tolerance. ABF3 in transgenic rice increased tolerance to drought stress alone. By using the 60 K Rice Whole Genome Microarray and RNA gel-blot analyses, we identified 12 and 7 target genes that were activated in transgenic rice plants by CBF3 and ABF3, respectively, which appear to render the corresponding plants acclimated for stress conditions. The target genes together with 13 and 27 additional genes are induced further upon exposure to drought stress, consequently making the transgenic plants more tolerant to stress conditions. Interestingly, our transgenic plants exhibited neither growth inhibition nor visible phenotypic alterations despite constitutive expression of the CBF3 or ABF3, unlike the results previously obtained from Arabidopsis where transgenic plants were stunted.
DOI:10.1016/j.plaphy.2014.03.030URLPMID:24751398 [本文引用: 1]

Various abiotic stresses downgrade the quality and productivity of chrysanthemum. A construct carrying both CcSOS1 (from Chrysanthemum crassum) and CdICE1 (from Chrysanthemum dichrum) was constitutively expressed in the chrysanthemum variety 'Jinba'. The transgenic plants were superior to the wild type (WT) ones with respect to their sensitivity to low temperature, drought and salinity, as measured by visible damage and plant survival. Salinity stressed transgenic plants accumulated more proline, and their level of superoxide dismutase and peroxidase activity was higher than in WT plants. At the physiological level, they suffered less loss of viable leaf area, maintained a lower leaf electrolyte conductivity and retained more chlorophyll (a+b). The ratio between the K(+) and Na(+) content was higher in the root, stem and median leaves of salinity stressed transgenic plants than in those of WT plants.
DOI:10.1007/s11427-006-2014-1URLPMID:17172050 [本文引用: 1]

DNA cassette containing an AtDREB1A cDNA and a nos terminator, driven by a cauliflower mosaic 35S promoter, or a stress-inducible rd29A promoter, was transformed into the ground cover chrysanthemum (Dendranthema grandiflorum) 'Fall Color' genome. Compared with wild type plants, severe growth retardation was observed in 35S:DREB1A plants, but not in rd29A:DREB1A plants. RT-PCR analysis revealed that, under stress conditions, the DREB1A gene was over-expressed constitutively in 35S:DREB1A plants, but was over-expressed inductively in rd29A:DREB1A plants. The transgenic plants exhibited tolerance to drought and salt stress, and the tolerance was significantly stronger in rd29A:DREB1A plants than in 35S:DREB1A plants. Proline content and SOD activity were increased inductively in rd29A:DREB1A plants than in 35S:DREB1A plants under stress conditions. These results indicate that heterologous AtDREB1A can confer drought and salt tolerance in transgenic chrysanthemum, and improvement of the stress tolerance may be related to enhancement of proline content and SOD activity.
DOI:10.1094/MPMI.2004.17.10.1162URLPMID:15497409 [本文引用: 1]

A new member of the tobacco (Nicotiana tabacum) AP2/ERF (ethylene response factor) transcription factor family, designated NtERF5, has been isolated by yeast one-hybrid screening. In vitro, recombinant NtERF5 protein weakly binds GCC box cis-elements, which mediate pathogen-regulated transcription of several PR (pathogenesis related) genes. NtERF5 transcription is transiently activated by wounding, by infection with the bacterial pathogen Pseudomonas syringae, as well as by inoculation with Tobacco mosaic virus (TMV). In contrast, NtERF5 transcription is not enhanced after application of salicylic acid, jasmonic acid, or ethylene. Constitutive overexpression of NtERF5 (ERF5-Oex) under control of the 35S promoter results in no visible alterations in plant growth or enhanced resistance to Pseudomonas infection. Furthermore, no constitutive expression of PR genes has been observed. In contrast, ERF5-Oex plants show enhanced resistance to TMV with reference to reduced size of local hypersensitive-response lesions and impaired systemic spread of the virus. Since, in TMV-infected ERF5-Oex plants, the viral RNA accumulates only up to 10 to 30% of the wild-type level, we suggest that NtERF5-regulated gene expression is controlling resistance to viral propagation. Previous research has demonstrated that overexpression of ERF genes enhances resistance to bacterial and fungal pathogens. Here, we provide further evidence that resistance to viral infection can be engineered by overexpression of ERF transcription factors.
DOI:10.1016/j.gene.2006.12.019URLPMID:17321073 [本文引用: 1]

ERF transcription factors can bind GCC boxes or non-GCC cis elements to regulate biotic and abiotic stress responses. Here, we report that an ERF transcription factor gene (GbERF2) was cloned by suppression subtraction hybridization from sea-island cotton after Verticillium dahliae attack. The GbERF2 cDNA has a total length of 1143 bp with an open reading frame of 597 bp. The genomic sequence of GbERF2 contains an intron of 515 bp. The gene encodes a predicated polypeptide of 198 amino acids with a molecular weight of 22.5 kDa and a calculated pI of 9.82. The GbERF2 protein has a highly conserved ERF domain while the nucleotide and amino acid sequences have low homology with other ERF plant proteins. An RNA blot revealed that GbERF2 is constitutively expressed in different tissues, but is higher in the leaves. High levels of GbERF2 transcripts rapidly accumulated when the plants were exposed to exogenous ethylene treatment and V. dahliae infection, while there was only a slight accumulation in response to salt, cold, drought and water stresses. In contrast, GbERF2 transcripts declined in response to exogenous abscisic acid (ABA) treatment. GbERF2 transgenic tobacco plants constitutively accumulated higher levels of pathogenesis-related gene transcripts, such as PR-1b, PR2 and PR4. The resistance of transgenic tobacco to fungal infection by Alternaria longipes was enhanced. However, the resistance to bacterial infection by Pseudomonas syringae pv. tabaci was not improved. These results show that GbERF2 plays an important role in response to ethylene stress and fungal attack in cotton.
DOI:10.1105/tpc.17.00349URLPMID:28550149 [本文引用: 1]

The plant hormone ethylene is critical for ripening in climacteric fruits, including apple (Malus domestica). Jasmonate (JA) promotes ethylene biosynthesis in apple fruit, but the underlying molecular mechanism is unclear. Here, we found that JA-induced ethylene production in apple fruit is dependent on the expression of MdACS1, an ACC synthase gene involved in ethylene biosynthesis. The expression of MdMYC2, encoding a transcription factor involved in the JA signaling pathway, was enhanced by MeJA treatment in apple fruits, and MdMYC2 directly bound to the promoters of both MdACS1 and the ACC oxidase gene MdACO1 and enhanced their transcription. Furthermore, MdMYC2 bound to the promoter of MdERF3, encoding a transcription factor involved in the ethylene-signaling pathway, thereby activating MdACS1 transcription. We also found that MdMYC2 interacted with MdERF2, a suppressor of MdERF3 and MdACS1 This protein interaction prevented MdERF2 from interacting with MdERF3 and from binding to the MdACS1 promoter, leading to increased transcription of MdACS1 Collectively, these results indicate that JA promotes ethylene biosynthesis through the regulation of MdERFs and ethylene biosynthetic genes by MdMYC2.
DOI:10.1104/pp.15.00677URLPMID:26103991 [本文引用: 1]

Ethylene is essential for many developmental processes and a key mediator of biotic and abiotic stress responses in plants. The ethylene signaling and response pathway includes Ethylene Response Factors (ERFs), which belong to the transcription factor family APETALA2/ERF. It is well known that ERFs regulate molecular response to pathogen attack by binding to sequences containing AGCCGCC motifs (the GCC box), a cis-acting element. However, recent studies suggest that several ERFs also bind to dehydration-responsive elements and act as a key regulatory hub in plant responses to abiotic stresses. Here, we review some of the recent advances in our understanding of the ethylene signaling and response pathway, with emphasis on ERFs and their role in hormone cross talk and redox signaling under abiotic stresses. We conclude that ERFs act as a key regulatory hub, integrating ethylene, abscisic acid, jasmonate, and redox signaling in the plant response to a number of abiotic stresses.
