 ,, 余爱丽
,, 余爱丽 ,山西省农业科学院谷子研究所/特色杂粮种质资源发掘与育种山西省重点实验室,山西长治 046011
,山西省农业科学院谷子研究所/特色杂粮种质资源发掘与育种山西省重点实验室,山西长治 046011Identification NADP-ME Gene of Foxtail Millet and Its Response to Stress
ZHAO JinFeng, DU YanWei, WANG GaoHong, LI YanFang, ZHAO GenYou, WANG ZhenHua, CHENG Kai, WANG YuWen ,, YU AiLi
,, YU AiLi ,Millet Research Institute, Shanxi Academy of Agricultural Sciences/Shanxi Key Laboratory of Genetic Resources and Breeding in Minor Crops, Changzhi 046011, Shanxi
,Millet Research Institute, Shanxi Academy of Agricultural Sciences/Shanxi Key Laboratory of Genetic Resources and Breeding in Minor Crops, Changzhi 046011, Shanxi通讯作者:
责任编辑: 李莉
收稿日期:2019-07-3接受日期:2019-09-22网络出版日期:2019-11-16
| 基金资助: |
Received:2019-07-3Accepted:2019-09-22Online:2019-11-16
作者简介 About authors
赵晋锋,Tel:0355-2204195;E-mail:zhaojfmail@126.com

摘要
关键词:
Abstract
Keywords:
PDF (6457KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵晋锋, 杜艳伟, 王高鸿, 李颜方, 赵根有, 王振华, 成凯, 王玉文, 余爱丽. 谷子NADP-ME的鉴定及其对逆境胁迫的响应[J]. 中国农业科学, 2019, 52(22): 3950-3963 doi:10.3864/j.issn.0578-1752.2019.22.002
ZHAO JinFeng, DU YanWei, WANG GaoHong, LI YanFang, ZHAO GenYou, WANG ZhenHua, CHENG Kai, WANG YuWen, YU AiLi.
0 引言
【研究意义】谷子(Setaria italica)是C4禾本科作物,具有抗旱、耐瘠、适应性广等特点,是起源于中国的重要粮食作物和饲草作物,在全球一些温带、亚热带和热带的干旱和半干旱地区广泛种植[1,2,3]。NADP-苹果酸酶(NADP-malic enzyme,NADP-ME)是一类广泛存在于植物界的氧化脱羧酶,在稳定细胞质pH和渗透势方面起重要作用[4,5],同时NADP-ME还参与多种胁迫响应,在植物应对干旱、高盐和高渗透等逆境胁迫信号途径中起重要作用[6]。【前人研究进展】植物体内NADP-ME分为光合型NADP-ME和非光合型NADP-ME [7]。光合型NADP-ME常定位于C4植物维管束鞘细胞叶绿体中或CAM植物的细胞质中,能催化苹果酸进行氧化脱羧反应,提供CO2用于光合作用过程中Rubisco酶的碳同化固定[8,9]。非光合型NADP-ME常定位于植物质体或细胞质中,在木质素、脂质合成,维持稳定细胞施渗透压、细胞质pH以及植物防御应答方面起重要作用[10,11,12,13,14]。NADP-ME通过催化苹果酸的代谢途径来参与植物体内多种生理反应,近年来,NADP-ME参与干旱、盐、低温等多种逆境胁迫的研究相继被报道,证明在逆境胁迫下植物体内NADP-ME代谢发生变化,相应酶活性提高,基因表达量增加,并在一定程度上提高了植物的耐逆能力[7]。在干旱胁迫条件下,水稻中3个NADP-ME表达量有不同程度的增加[15],烟草进行干旱胁迫后叶片中NADP-ME活性增加了3.9倍,定量PCR检测显示NADP-ME的mRNA表达量及酶活性均有所增加[16]。研究表明干旱会导致植物叶表皮保卫细胞ABA浓度增加,进而引起NADP-ME酶积累,苹果酸代谢活跃含量下降,因此,推测NADP-ME可能通过ABA介导激活对干旱胁迫响应的信号途径[17]。研究表明水稻幼苗中NADP-ME的表达在氯化钠和碳酸盐处理下受到强烈的诱导,酶活性也明显增加,该基因在拟南芥中过表达后耐逆性明显增强[18,19]。毛果杨中5个NADP-ME在氯化钠胁迫下表达均受到不同程度地诱导[20]。NADP-ME是定位于膜上的亲水性蛋白,低温胁迫可以诱导NADP-ME转录增强,在小麦和巴西橡胶树中都得到了验证[20,21]。【本研究切入点】谷子基因组测序已完成并公布[22,23],但谷子NADP-ME家族基因的鉴定以及NADP-ME在非生物逆境胁迫下的表达分析尚未见报道。【拟解决的关键问题】本研究鉴定谷子基因组中NADP-ME基因,明确家族基因的结构特点与进化特征,通过实时荧光定量PCR技术分析该基因家族成员在苗期不同逆境、不同生育期下干旱胁迫和不同光照强度下的动态表达特征及差异,为进一步研究NADP-ME基因在逆境应答过程中的作用奠定基础。1 材料与方法
1.1 试材与处理
试验于2018年在山西省农业科学院谷子研究所进行。所用试验材料为谷子品种豫谷1号。2018年5月在组培室和旱棚分别种植材料,待组培室幼苗长至三叶一心期时分别对其进行20% PEG6000模拟干旱、盐(250 mmol·L-1 NaCl)、ABA(100 μmol·L-1)和低温(4℃)胁迫处理,设立时间点0、1、3、6、12和24 h整株取样[24]。另外在旱棚种植豫谷1号,对照生育期内正常浇水,浇水时间点选择具体措施为观察旱棚对照小区内土壤含水量呈明显降低,植株叶片颜色呈现鲜绿逐渐丧失开始向灰绿转变但叶片尚未卷曲时马上浇透水。对照小区整个生育期内共浇水9次;干旱处理小区只浇3次关键水,分别为拔节(6月20日)、抽穗(8月5日)和灌浆期(8月30日)浇透水,其他时间采用自然控水方式控水。对照小区和干旱处理小区施肥、中耕等其他农田管理措施完全相同。干旱处理不同生育期(拔节、抽穗、灌浆)叶片样本在对应生育期浇水前与对照同时取样。不同光照处理为当植株出苗后用黑色遮阳网分别遮挡1层(中等光照Ⅰ,拔节期、抽穗期、灌浆期测量光照强度分别为1.32×104、2.45×10 4和0.98×10 4 lx)、2层(弱光照Ⅱ,拔节期、抽穗期、灌浆期测量光照强度分别为0.31×10 4、0.57×10 4和0.35×10 4 lx)至成熟收获。使用标智光强测定仪(GM1040)于晴天上午9:00—10:00点测量光照强度,测量10次数值,取平均值。旱棚材料分别在拔节、抽穗、灌浆期时取对照和处理旗叶叶片,所有样品叶片取样后立即在-80℃冰箱中速冻备用。1.2 总RNA提取、cDNA合成及引物设计
使用生工TRIzol试剂盒提取植物总RNA;使用生工第一链cDNA合成试剂盒(RevertAid Premium Reverse Transcriptase)合成cDNA,上述试验均按照试剂盒说明书进行。用Primer Primer 5.0软件设计引物。1.3 基因发掘和生物信息学分析
根据文献获得已知玉米NADP-ME蛋白序列[25],将获得的蛋白序列提交到谷子基因组数据库(https://genome.jgi.doe.gov)搜索可能的NADP-ME同源基因,然后用InterProScan搜索候选谷子NADP-ME基因的结构特征域,把同时含NADP依赖性苹果酸酶(PLN03129)、苹果酸酶(SfcA)、苹果酸酶N端功能域(PF00390)和苹果酸酶NAD结合功能域(PF03949)的谷子同源NADP-ME基因确定为谷子候选NADP-ME基因。通过谷子数据库网站(https://genome.jgi.doe.gov)检索基因序列号、基因组序列长度、CDS长度、氨基酸数目等参数;利用ExPASy网站(https://prosite.expasy.org)在线工具预测蛋白分子量(MW)和等电点(pI)。利用在线软件plantCARE (http://bioinformatics.psb.ugent.be/ webtools/plantcare/html/)分析启动子顺式元件,采用GSDS软件(http://gsds.cbi.pku.edu.cn/)绘制基因结构图,用Psort在线工具预测亚细胞定位,使用BLAST工具在NCBI上查找氨基酸同源性序列,用ClustalX1.83软件进行基因序列比对[26];使用Mega6.0软件邻接法构建不同物种N-J系统进化树[27]。1.4 Real-time PCR分析
所用引物由Primer Primer 5.0设计,引物信息见表1。样品cDNA均一化后作为实时荧光定量PCR模板,以谷子β-actin(Seita. 7G294000)作为内参基因。反应体系为10 μL 2×荧光染料混合液(BBI, Shanghai, China)、0.4 μL正向引物(10 μmol·L-1)、0.4 μL反向引物(10 μmol·L-1)、2 μL cDNA模板和7.2 μL无菌水。经预试验优化后,PCR反应程序为95℃ 3 min;95℃ 7 s,57℃ 10 s,72℃ 15 s,45个循环。试验设计3次重复,采用相对定量2-ΔΔCt方法计算基因在某种逆境处理下某个时间点相对于对照的转录水平变化[28]。每个处理3次重复。采用2-ΔΔCt法计算相对表达量,采用SPSS19.0软件对数据进行统计分析。苗期不同逆境下表达分析结果利用百迈克云平台在线工具(https://international.biocloud.net/zh/software/tools/detail/small/305)聚类热图绘制(Pretty Heatmaps)进行热图绘制,参数默认。Table 1
表1
表1本试验所用引物
Table 1
| 基因 Gene | 正向引物 Forward primer (5′-3′) | 反向引物 Reverse primer (5′-3′) |
|---|---|---|
| SiNADP-ME1 | AGGAGCATCCAGGTCATCGT | ACTCGTCGTTGAGCAAGGTCT |
| SiNADP-ME2 | AAAATTTATGGTAGATGTCAGGTCC | TGACTACCTTCTGCCAAATCCT |
| SiNADP-ME3 | TTACACACCGACTGTTGACGAG | TGAGCACCCAAATCTCCAAGT |
| SiNADP-ME4 | CTCCGATTGAAGAGTGCCG | TGTGAATGCCCTTCCAACTC |
| SiNADP-ME5 | GCAAGATATGGCACAACTCACC | CCAGCACCCAGGAATAGGAA |
| SiNADP-ME6 | ACCTATCATCCTCGCTCTATCAA | CAAATGGGCTTCCACTTCC |
| SiNADP-ME7 | AAATTTTATGGTAGATGTCAGGTCC | TTTCATCCACTGGTGAGAACTTC |
| β-Actin | GGGCAGTTGCTATCCACATC | ATCACGCCACAAGACCAGAC |
新窗口打开|下载CSV
2 结果
2.1 谷子NADP-ME基因家族成员的鉴定与生物信息学分析
2.1.1 谷子NADP-ME基因的鉴定 参考已知NADP- ME序列,在谷子全基因组中鉴定出7个SiNADP-ME候选基因成员(表2)。根据基因在基因组上的相应位置分别命名为SiNADP-ME1、SiNADP-ME2、SiNADP-ME3、SiNADP-ME4、SiNADP-ME5、SiNADP- ME6和SiNADP-ME7。7个基因不均分布在第2、3、5、7染色体上,其中第5染色体上有3个SiNADP-ME,第3染色体上有2个,第2和7染色体上各有1个基因。序列分析显示SiNADP-ME成员之间基因组长度变化明显,SiNADP-ME1、SiNADP-ME4、SiNADP-ME5、SiNADP-ME6基因组序列较长,分别含3 400、5 236、5 357和4 665个碱基;而SiNADP-ME2、SiNADP-ME3和SiNADP-ME7基因组序列较短,分别含2 128、2 329和2 446个碱基。GSDS在线软件基因结构分析表明,SiNADP-ME1、SiNADP-ME2、SiNADP-ME3和SiNADP-ME7内含子较少,分别为8、6、8和4个,而SiNADP-ME4、SiNADP-ME5和SiNADP-ME6内含子较多,分别为19、19和18个(图1)。另外发现SiNADP-ME2、SiNADP-ME3、SiNADP-ME4、SiNADP- ME6和SiNADP-ME7只有一种转录方式,而SiNADP- ME1有2个可变剪切,SiNADP-ME5有3个可变剪切,后续分析时将SiNADP-ME1和SiNADP-ME5同一基因位点出现的多个可变剪切体视为同一基因,仅以原始转录变异体(primary alternative transcript)对应的序列作为该基因的代表。Table 2
表2
表2预测谷子NADP-ME参数
Table 2
| 基因 Gene | 基因 ID Gene ID | 染色体位置 Chr. location | 基因长度 Genome length (bp) | 氨基酸 数目 Number of amino acid | 等电点 pI | 分子量 Molecular weight (kD) | 内含子数Intro number | 不稳定 指数Instability index | 脂肪系数Aliphatic index | 平均疏水指数 Grand average of hydropathicity |
|---|---|---|---|---|---|---|---|---|---|---|
| SiNADP-ME1 | Seita.3G109300 | 278989-7282388 | 3400 | 576 | 5.60 | 637.72 | 8 | 39.31 | 91.46 | -0.176 |
| SiNADP-ME2 | Seita.3G284800 | 26806748-26808875 | 2128 | 213 | 5.47 | 233.24 | 6 | 32.64 | 91.60 | -0.209 |
| SiNADP-ME3 | Seita.4G119800 | 12185662-12187990 | 2329 | 265 | 5.32 | 296.11 | 8 | 38.32 | 107.77 | 0.004 |
| NSiADP-ME4 | Seita.5G134300 | 11688238-11693473 | 5236 | 639 | 6.31 | 700.38 | 19 | 45.01 | 89.19 | -0.138 |
| SiNADP-ME5 | Seita.5G301800 | 35587449-35592805 | 5357 | 652 | 6.28 | 725.43 | 19 | 44.52 | 89.49 | -0.218 |
| SiNADP-ME6 | Seita.5G314300 | 36545367-36550031 | 4665 | 636 | 8.05 | 698.51 | 18 | 42.98 | 92.25 | -0.133 |
| SiNADP-ME7 | Seita.7G040900 | 12423964-12426409 | 2446 | 149 | 5.40 | 161.94 | 4 | 23.01 | 97.52 | -0.054 |
新窗口打开|下载CSV
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1SiNADP-ME成员的基因结构
Fig. 1Gene structure of SiNADP-ME genes
2.1.2 SiNADP-ME蛋白特性分析 采用EXPASY网站在线工具对候选SiNADP-ME蛋白的分子量、等电点、不稳定系数、脂肪系数、平均疏水指数进行预测(表2),基因组序列较长的SiNADP-ME1、SiNADP-ME4、SiNADP-ME5和SiNADP-ME6分别编码576、639、652和636个氨基酸,而基因组序列较短的SiNADP-ME2、SiNADP-ME3和SiNADP- ME7分别编码213、265和149个氨基酸。参数预测结果显示SiNADP-ME成员间分子量跨度较大,范围在161.94—725.43 kD,蛋白等电点范围为5.32—8.05,不稳定指数范围为23.01—45.01,脂肪系数范围为89.19—107.77,平均疏水指数范围为-0.218—0.004。
2.1.3 SiNADP-ME蛋白亚细胞定位预测和启动子区域顺式元件分析 采用Psort在线工具对谷子SiNADP-ME家族成员的亚细胞定位进行预测,结果表明,SiNADP-ME成员主要被定为在叶绿体、线粒体和细胞质中,另外也可能存在于细胞外、细胞核和过氧化物酶体中(表3)。利用plantCARE在线软件对SiNADP-ME成员启动子顺式元件进行分析(表4),SiNADP-ME成员启动子区域主要包括激素类应答元件(脱落酸、水杨酸、茉莉酸甲酯、赤霉素)、逆境应答元件(干旱、低温、防御和应激反应)、大量的光应答元件以及其他类生长调控相关顺式元件,包括缺氧特异性诱导、厌氧诱导必需、分生组织特异性表达、玉米蛋白代谢调控、种子特异性调控、细胞周期调控、昼夜节律元件等。
Table 3
表3
表3SiNADP-ME蛋白亚细胞定位预测
Table 3
| 基因 Gene | 预测位置 Localization | 可信度 Reliability |
|---|---|---|
| SiNADP-ME1 | 细胞质Cytoplasmic ;叶绿体Chloroplast ;线粒体Mitochondrial | 2.254;1.083;0.957 |
| SiNADP-ME2 | 细胞质Cytoplasmic ;细胞外Extracellular ; 细胞核Nuclear | 3.030;0.452;0.401 |
| SiNADP-ME3 | 细胞质Cytoplasmic ;线粒体Mitochondrial ;叶绿体Chloroplast | 1.834;0.753;0.752 |
| SiNADP-ME4 | 叶绿体Chloroplast ;线粒体Mitochondrial ;细胞质Cytoplasmic | 2.491;1.274;0.390 |
| SiNADP-ME5 | 线粒体Mitochondrial ;叶绿体Chloroplast ;过氧化物酶体Peroxisomal | 1.463;1.380;0.829 |
| SiNADP-ME6 | 线粒体Mitochondrial ;叶绿体Chloroplast ;过氧化物酶体Peroxisomal | 2.206;1.616;0.449 |
| SiNADP-ME7 | 细胞质Cytoplasmic;细胞外Extracellular;细胞核Nuclear | 1.351;1.144;0.598 |
新窗口打开|下载CSV
Table 4
表4
表4SiNADP-ME启动子区域顺式元件预测
Table 4
| 顺式元件 Cis-element | 典型序列 Typical sequence | 特性 Characteristic | 基因 Gene |
|---|---|---|---|
| ABRE | ACGTG | 脱落酸响应 Abscisic acid responsiveness | SiNADP-ME1,2,4,5,6,7 |
| CACGTG | SiNADP-ME2,4,6 | ||
| GCCGCGTGGC | SiNADP-ME1,4,6 | ||
| TCA-element | TCAGAAGAGG | 水杨酸响应 Salicylic acid responsiveness | SiNADP-ME4,7 |
| CGTCA-motif | CGTCA | 茉莉酸甲酯响应 MeJA-responsiveness | SiNADP-ME1,3,4,5,6,7, |
| TGACG-motif | TGACG | SiNADP-ME3,4,5,6,7 | |
| P-box | CCTTTTG | 赤霉素响应 Gibberellin-responsive element | SiNADP-ME1,3,6 |
| GARE-motif | TCTGTTG | SiNADP-ME3, | |
| TATC-box | TATCCCA | SiNADP-ME6 | |
| TGA-element | AACGAC | 生长素响应 Auxin-responsive element | SiNADP-ME1,5 |
| LTR | CCGAAA | 低温响应 Low-temperature responsiveness | SiNADP-ME4,5 |
| MBS | CAACTG | 干旱响应 Drought-inducibility | SiNADP-ME3,4,5,,7 |
| TC-rich repeats | GTTTTCTTAC | 防御和应激反应响应 Defense and stress responsiveness | SiNADP-ME6 |
| G-box | CACGTT | 光响应 Light responsiveness | SiNADP-ME4,5,7 |
| CACGTG | SiNADP-ME2,4,6 | ||
| CACGTC | SiNADP-ME1,2,4 | ||
| TACGTG | SiNADP-ME1,2,6 | ||
| Sp1 | GGGCGG | 光响应 Light responsive element | SiNADP-ME2,6,7 |
| GT1-motif | GGTTAA | SiNADP-ME1,2,4,5 | |
| 3-AF1 binding site | TAAGAGAGGAA | SiNADP-ME2,3 | |
| TCCC-motif | TCTCCCT | 光响应 Part of a light responsive element | SiNADP-ME2,4,7 |
| GATA-motif | AAGGATAAGG | SiNADP-ME2,3 | |
| GATAGGG | SiNADP-ME5,6 | ||
| GC-motif | CCCCCG | 缺氧特异性诱导 Anoxic specific inducibility | SiNADP-ME1,2,4,7 |
| CAT-box | GCCACT | 分生组织响应 Meristem expression | SiNADP-ME1,2,4,6,7 |
| ARE | AAACCA | 厌氧响应 Essential for the anaerobic induction | SiNADP-ME4,5 |
| O2-site | GATGACATGG | 玉米蛋白代谢调控 Zein metabolism regulation | SiNADP-ME1,4,5 |
| CCAAT-box | CAACGG | MYBHv1结合位点 MYBHv1 binding site | SiNADP-ME1,5 |
| RY-element | CATGCATG | 种子特异性调控 Seed-specific regulation | SiNADP-ME1,4 |
| MSA-like | (T/C)C(T/C)AACGG(T/C)(T/C)A | 细胞周期调控 Cell cycle regulation | SiNADP-ME1 |
| circadian | CAAAGATATC | 昼夜节律 Circadian control | SiNADP-ME3 |
| motif I | gGTACGTGGCG | 根特异性 Root specific | SiNADP-ME6 |
新窗口打开|下载CSV
2.1.4 NADP-ME序列比对及进化分析 ClustalX1.83软件比对谷子NADP-ME成员氨基酸序列发现成员之间序列非常保守,相似性较较高,所有SiNADP-ME序列一致性(Identity)为54.43%(图2)。与SiNADP-ME1、SiNADP-ME4、SiNADP-ME5和SiNADP- ME6序列相比,SiNADP-ME2、SiNADP-ME3和SiNADP-ME7序列缺少一段N端序列,但是它们都含有NADP依赖性苹果酸酶(PLN03129)、苹果酸酶(SfcA)、苹果酸酶N端功能域(PF00390)和苹果酸酶NAD结合功能域(PF03949)等NADP基因典型功能域。序列比对发现SiNADP-ME1、SiNADP-ME4、SiNADP-ME5和SiNADP-ME6之间同源性较高,序列一致性为77.30%。为进一步了解谷子SiNADP-ME进化关系,推测其生物功能,在NCBI数据库中检索了不同物种NADP-ME基因,包括玉米、高粱、水稻、小麦、拟南芥以及北美云杉和小立碗藓(玉米ZmNADP-ME1、ZmNADP-ME2高粱SbNADP-ME1、SbNADP-ME2,水稻OsNADP-ME1、OsNADP-ME2,小麦TaNADP-ME1、TaNADP-ME2,拟南芥AtNADP- M1、AtNADP-M2,北美云杉PtNADP-ME1、PtNADP- ME2,小立碗藓PpNADP-ME1、PpNADP-ME2,对应GenBank序列号分别为NP_001150965、XP_008656303,XP_002440734、XP_021312641,BAA03949、XP_ 015640686,ABW77317、CDM81737,NP_197960、NP_178093,AEX13395、AEX13397,PNR56966,XP_024371400)。利用MEGA6.0软件构建了谷子和不同物种NADP-ME蛋白成员的Neighbor-Joining进化树(图3)。从图3可以看出,所有物种NADP-ME序列非常保守,其序列一致性为56.52%。谷子的4个基因SiNADP-ME2、SiNADP-ME3、SiNADP-ME4和SiNADP-ME7聚合在一起以基因簇的方式出现,揭示它们在进化上可能由共同祖先复制而来,另一方面也暗示它们在某些功能上可能具有相似性。玉米、高粱、水稻、小麦、拟南芥NADP-ME成员与谷子NADP-ME成员相互嵌合在一起,表明植物NADP-ME基因在进化过程中相对保守。另外发现一些同源基因对,例如SiNADP-ME1和OsNADP-ME2,NADP-ME5、SbNADP- ME2和ZmNADP-ME1,SbNADP-ME1和ZmNADP- ME2,这些物种的NADP-ME聚在一起表明它们的亲缘关系较近,推测这些NADP-ME对可能具有类似的生物功能。小立碗藓的2个PtNADP-ME单独聚在一起,表明与其他物种NADP-ME成员间的亲缘关系较远,揭示PtNADP-ME可能与其他物种NADP-ME成员之间的生物功能差异较大。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2NADP-ME家族蛋白序列的氨基酸聚类分析
OsNADP-ME2:水稻Oryza sativa,XP_015640686;TaNADP-ME1:小麦Triticum aestivum,ABW77317;ZmNADP-ME2:玉米Zea mays,XP_008656303;SbNADP-ME1:高粱Sorghum bicolor,XP_002440734;ZmNADP-ME1:玉米Zea mays,NP_001150965;OsNADP-ME1:水稻Oryza sativa,BAA03949;TaNADP-ME2:小麦Triticum aestivum,CDM81737;AtNADP-M2:拟南芥Arabidopsis thaliana,NP_178093;AtNADP-M1:拟南芥Arabidopsis thaliana,NP_197960;SbNADP-ME2:高粱Sorghum bicolor,XP_021312641;PpNADP-ME1:小立碗藓Physcomitrella patens,PNR56966;PpNADP-ME2:小立碗藓Physcomitrella patens,XP_024371400;PtNADP-ME1:北美云杉Picea sitchensis,AEX13395;PtNADP-ME2:北美云杉Picea sitchensis,AEX13397。相同氨基酸残基用黑色表示,相似氨基酸残基用灰色表示(≥60% similarity)The amino acids with an entire homology are shown by a black background, and those shared non-identical conserved identity by a gray background (≥ 60% similarity)
Fig. 2Amino acid sequence alignment of NADP-MEs
OsNADP-ME2:水稻Oryza sativa,XP_015640686;TaNADP-ME1:小麦Triticum aestivum,ABW77317;ZmNADP-ME2:玉米Zea mays,XP_008656303;SbNADP-ME1:高粱Sorghum bicolor,XP_002440734;ZmNADP-ME1:玉米Zea mays,NP_001150965;OsNADP-ME1:水稻Oryza sativa,BAA03949;TaNADP-ME2:小麦Triticum aestivum,CDM81737;AtNADP-M2:拟南芥Arabidopsis thaliana,NP_178093;AtNADP-M1:拟南芥Arabidopsis thaliana,NP_197960;SbNADP-ME2:高粱Sorghum bicolor,XP_021312641;PpNADP-ME1:小立碗藓Physcomitrella patens,PNR56966;PpNADP-ME2:小立碗藓Physcomitrella patens,XP_024371400;PtNADP-ME1:北美云杉Picea sitchensis,AEX13395;PtNADP-ME2:北美云杉Picea sitchensis,AEX13397。The amino acids with an entire homology are shown by a black background, and those shared non-identical conserved identity by a gray background (≥ 60% similarity)
图3
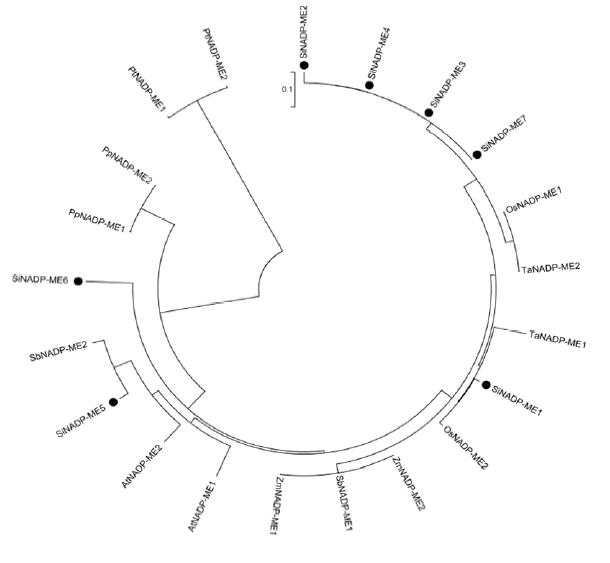 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3不同物种NADP-ME蛋白的进化关系
Fig. 3Phylogenetic relationships of NADP-ME proteins from different species
2.2 SiNADP-ME家族基因的表达分析
2.2.1 SiNADP-ME非生物逆境胁迫下表达分析 如图4所示,谷子三叶一心期幼苗4种胁迫处理下7个SiNADP-ME基因在不同时间点表达水平变化趋势不完全相同。除了SiNADP-ME2和SiNADP-ME7在PEG胁迫处理下所有时间点表达量下调外,所有基因表达量在4种胁迫处理过程中至少1个时间点均有所上调。另外发现低温胁迫下,所有SiNADP-ME的相对表达量都在胁迫后24 h达到最高分别为对照的411.50、12.36、3.49、3.36、15.90、15.21和10.92倍。SiNADP-ME1在ABA处理3 h、低温处理24 h、NaCl处理12 h后,它们的相对表达量均达到最高,分别为对照的460.53、411.50和15.24倍;SiNADP-ME6在ABA处理12 h、低温处理24 h、PEG处理12 h、NaCl处理24 h后,它们的相对表达量均达到最高,分别为对照的211.13、15.21、772.41和643.99倍;除此之外其他基因在其他的逆境胁迫下的表达量上调范围均在对照的1—5倍。图4
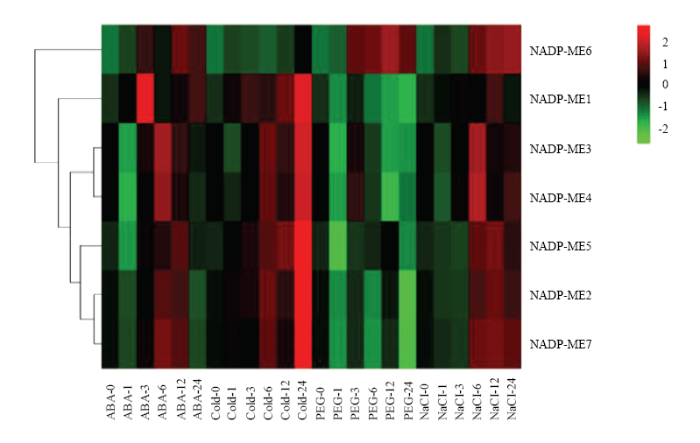 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4谷子SiNADP-ME表达谱
图中浅绿、深绿、黑色、浅红、深红五色代表基因表达水平。绿色表示基因表达弱,红色表示基因表达强
Fig. 4Expression profile of SiNADP-ME gene in foxtail millet
Light green,dark green, black, light red and dark red are used to represent gene expression levels. Green indicates weak gene expression, red indicates strong gene expression
2.2.2 不同生育期干旱胁迫及不同光照强度下表达分析 为进一步了解NADP-ME在谷子不同生育期干旱胁迫下的表达情况,选取SiNADP-ME1和SiNADP-ME6 2个基因检测了它们在谷子拔节期、抽穗期、灌浆期3个关键生育期干旱胁迫及不同光照强度下的表达情况(图5),SiNADP-ME1在正常抽穗期、灌浆期表达量与拔节期相比均有明显提升,分别为拔节期的1.72和12.07倍;在干旱胁迫下的抽穗期、灌浆期表达量与拔节期相比均也有明显提升,分别为拔节期的2.02和1.84倍。SiNADP-ME6在正常与干旱条件下的灌浆期、抽穗期表达量与拔节期相比也均有明显提升。结果表明,SiNADP-ME1和SiNADP-ME6在正常生长条件下随着谷子生长发育其表达有所增强,在不同生育期干旱胁迫下其表达量都有明显增加,表明2个基因都参与了对干旱胁迫下的响应。如图6所示,SiNADP-ME1在拔节期和抽穗期干旱条件下表达量较对照均有明显提高,但灌浆期却有所下降,而SiNADP-ME6在抽穗期和灌浆期干旱条件下表达量较对照有明显提高,但拔节期却有所下降。表明SiNADP-ME1和SiNADP-ME6在干旱条件下不同生育期表达特征有所不同。不同光照强度下,SiNADP-ME1在拔节期弱光(光照Ⅱ)下表达量较高,为拔节期正常光照的3.50倍,而在拔节期中等光照(光照Ⅰ)及抽穗和灌浆期的中等光照和弱光照下表达量都比较低。SiNADP-ME6在各个生育期拔节期中等光照下表达量都比较低,在拔节期和抽穗期的低光照强度下甚至检测不到信号。结果表明,光照强度的强弱严重影响SiNADP-ME1和SiNADP-ME6的表达。
图5
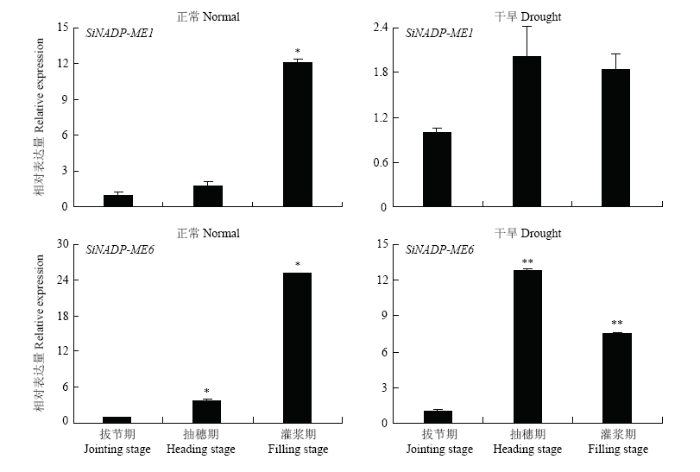 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5正常和干旱条件下SiNADP-ME1和SiNADP-ME6 在不同生育期的相对表达量
*表示在0.05水平上显著,**表示在0.01水平上显著。下同
Fig. 5Relative expression of SiNADP-ME1 and SiNADP-ME6 under normal and drought conditions during different growth stages
Single asterisk show the discrepancy on 0.05 levels notable and double asterisk show the discrepancy on 0.01 levels notable. The same as below
图6
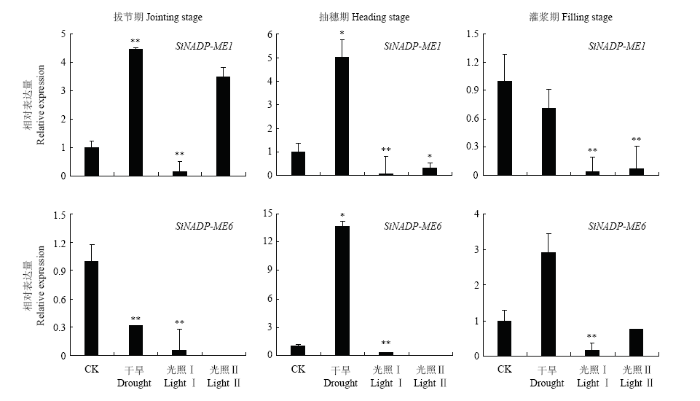 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6光照和干旱条件下SiNADP-ME1和SiNADP-ME6 的相对表达量
Fig. 6Relative expression of SiNADP-ME1 and SiNADP-ME6 under light and drought conditions
3 讨论
本研究通过生物信息学方法在谷子基因组中检索得到7个NADP-ME基因,集中分布在谷子的4条染色体上(2、3、5、7)。其中SiNADP-ME1和SiNADP-ME5具有可变剪切,其他5个基因只有一种转录方式。7个基因中4个基因序列较长(SiNADP-ME1、SiNADP-ME4、SiNADP-ME5和SiNADP-ME6),3个基因由于缺失了N端部分序列而较短(SiNADP-ME2、SiNADP-ME3、SiNADP-ME7),但是NCBI保守功能域分析显示7个基因蛋白都含有NADP-ME保守特征功能域,因此推断这7个基因是谷子NADP-ME家族成员。采用ClustalX1.83比较不同物种中NADP-ME蛋白序列,发现所有NADP-ME基因序列同源性较高,具有非常保守的序列和结构(图2)。系统进化树表明谷子NADP-ME与高粱、玉米、水稻、小麦NADP-ME紧密嵌合在一起,而与拟南芥、北美云杉和小立碗藓相距较远(图3),揭示了谷子与高粱、玉米、水稻亲缘关系相对较近,而与拟南芥、北美云杉和小立碗藓亲缘关系较远。这与它们在植物分类上的亲缘关系远近是一致的(谷子、高粱、玉米同属被子植物禾本科单子叶C4植物,水稻属被子植物禾本科单子叶作物,拟南芥属被子植物十字花科双子叶植物,北美云杉属裸子植物门松科植物,小立碗藓属苔藓植物门葫芦藓科)。系统发育进化树中谷子NADP-ME分别和单子叶作物高粱、玉米、水稻、小麦以及双子叶植物拟南芥相应基因聚在一起说明谷子NADP-ME在单双子叶植物分离之前就已存在。另外在进化树中发现一些不同物种构成的同源基因对,揭示这些同源基因对可能由共同祖先进化而来,另一方面也暗示它们在某些信号通路中可能具有相似的功能。此外对谷子7个SiNADP-ME基因生物信息学特征进行了系统的预测和分析,期望能够发现功能研究有帮助信息。预测结果显示SiNADP-ME很多性状和参数非常接近类似,比如亚细胞定位分析SiNADP-ME成员主要被定为在细胞的叶绿体、线粒体和细胞质中,另外也可能存在于细胞外、细胞核和过氧化物酶体中,这些位置都是植物光合作用的关键位置,揭示了SiNADP-ME可能在植物光合作用信号途径中起比较重要作用。这些结论不仅验证了它们同属NADP-ME基因家族,而且预示不同NADP-ME可能共同参与或调控某些信号途径。顺式元件分析表明在SiNADP-MEs启动子区域含有ABA、低温、干旱和防御应激反应等顺式元件。一般来讲如果基因启动子区域存在某种顺式元件则暗示该基因很可能参与相应的信号途径[29]。研究表明在非生物逆境,比如干旱、盐、低温、高温以及伤害等胁迫下会导致植物细胞内的ABA水平升高,而且众所周知ABA是在非生物逆境应答中起重要作用的激素[30,31,32]。因此推测谷子SiNADP-ME很可能参与了植物对非生物逆境胁迫的响应。所以用Real-time PCR检测SiNADP-ME在谷子幼苗期不同逆境胁迫下的表达情况,结果表明,7个谷子SiNADP-ME基因在4种处理下其表达量有显著变化,但具体动态表达模式不尽相同。如图4所示,只有SiNADP-ME2和SiNADP-ME7在PEG胁迫处理下所有时间点表达量下调,其他所有SiNADP-ME的表达量在不同胁迫处理过程中至少1个时间点表现上调。另外7个SiNADP-ME在低温胁迫处理下相对表达量都在24 h达到最高,分别为对照的411.50、12.36、3.49、3.36、15.90、15.21和10.92倍。试验结果表明谷子SiNADP-ME家族基因广泛参与了植物非生物逆境胁迫应答。本研究结果同已有关于NADP-ME在逆境应答方面研究结果是相一致的。例如FU等[33]发现在PEG、低温(4℃)、黑暗、盐(200 mmol·L-1 NaCl)、脱落酸和水杨酸胁迫处理下,晋麦47中NADP-ME酶的活性有明显提高,进一步研究表明NADP-ME酶在小麦植株对各种逆境胁迫的反应中起重要作用。CUSHMAN等[34]发现盐处理下,冰草NADP-ME的mRNA转录水平增加8—10倍。芦芸经盐处理后,NADP-ME的转录水平在12 h被显著诱导,转录水平在24 h达到最大,NADP-ME酶活性一直持续增加到72 h[35]。CHI等[36]对水稻叶中4个NADP-ME基因的研究发现,3个基因(OsNADP-ME1、OsNADP-ME2和OsNADP-ME4)在ABA(50 μmol·L-1)和25% PEG8000处理下表达量增加。值得注意的是SiNADP-ME1和SiNADP-ME6,它们在某些逆境胁迫下被诱导表达量上调趋势非常之大,比如SiNADP-ME1在ABA处理3 h、低温处理24 h后它们的相对表达量均达到最高分别为对照的460.53和411.50倍;SiNADP-ME6在ABA处理12 h、PEG处理12 h、NaCl处理24 h后它们的相对表达量均达对照的211.13、772.41和643.99倍。因此推测SiNADP-ME1和SiNADP-ME6在可能逆境应答中发挥重要作用。为了进一步了解SiNADP-ME在不同关键生育期干旱胁迫下的表达情况,检测了SiNADP-ME1和SiNADP-ME6在不同生育期(拔节期、抽穗期和灌浆期)干旱胁迫及不同光照强度(中等光照强度Ⅰ和弱光照强度Ⅱ)下的表达情况下的表达情况。如图5所示,结果表明,在正常条件下SiNADP- ME1和SiNADP-ME6随着谷子生长发育营养生长和光合作用加强其表达显著提高,在不同生育期干旱胁迫下其表达量都有明显增加,说明2个基因在不同生育期都参与了对干旱胁迫下的响应。不同光照强度下,除了SiNADP-ME1在拔节期弱光下表达量较高外,2个基因在3个关键生育期的中等光照和弱光照下表达量都比较低,有的甚至检测不到信号(图6)。试验结果揭示SiNADP-ME1和SiNADP-ME6可能参与了谷子在拔节、抽穗、灌浆期的干旱应答,光照强度的强弱严重影响SiNADP-ME1和SiNADP-ME6在不同生育期的表达。这些试验结论进一步验证了顺式元件分析结果,证明了SiNADP-ME在植物逆境应答中起一定作用。顺式元件分析还显示在SiNADP-ME启动子区域存在茉莉酸甲酯、水杨酸、赤霉素顺式元件。研究表明病原体感染通常导致细胞中茉莉酸甲酯、水杨酸、乙烯等激素水平的增加[37],因此,推断SiNADP-ME可能在生物应激反应中起一些作用。此外还发现了缺氧特异性诱导(GC-motif)、分生组织表达(CAT-box)、厌氧诱导必需(ARE)、玉米醇溶蛋白代谢调控(O2-site)、种子特异性调控(RY-element)、细胞周期调控(MSA-like)、昼夜节律核心元件(circadian)、根特异性(motif I),这些顺式元件的存在暗示SiNADP-ME可能参与相应的生理生化过程。值得一提的是在启动子区域还发现了大量的光应答元件,众所周知谷子是光温敏感性作物,预示SiNADP-ME可能参与调控谷子的光温应答调控。尽管这些推测都需要严谨的试验证明,但还是为今后的功能研究提供了一些线索。本文报道的谷子SiNADP-ME丰富和完善了植物NADP-ME家族成员,为进一步阐明NADP-ME在谷子逆境应答中的功能、机制提供了试验依据。4 结论
从谷子全基因组中鉴定出7个NADP-ME成员,集中分布在谷子第2、3、5、7染色体上。所有成员蛋白序列高度保守,都具有NADP-ME保守特征功能域。谷子NADP-ME家族基因成员主要存在于叶绿体、线粒体和细胞质。SiNADP-ME广泛参与了谷子苗期非生物逆境胁迫应答,尤其是SiNADP-ME1和SiNADP-ME6参与了谷子在拔节、抽穗、灌浆期的干旱应答,而光照强度的强弱会严重影响SiNADP-ME1和SiNADP-ME6在不同生育期的表达。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
