 ,, 韩贝, Abid Ullah, 张献龙, 杨细燕*华中农业大学作物遗传改良国家重点实验室, 湖北武汉 430070
,, 韩贝, Abid Ullah, 张献龙, 杨细燕*华中农业大学作物遗传改良国家重点实验室, 湖北武汉 430070Fungi diversity analysis of rhizosphere under drought conditions in cotton
YUE Dan-Dan ,, HAN Bei, Abid Ullah, ZHANG Xian-Long, YANG Xi-Yan*National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China
,, HAN Bei, Abid Ullah, ZHANG Xian-Long, YANG Xi-Yan*National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China通讯作者: * 杨细燕, E-mail:yxy@mail.hzau.edu.cn
收稿日期:2020-07-20接受日期:2021-01-21网络出版日期:2021-02-25
| 基金资助: |
Corresponding authors: * E-mail:yxy@mail.hzau.edu.cn
Received:2020-07-20Accepted:2021-01-21Online:2021-02-25
| Fund supported: |
作者简介 About authors
E-mail:ddyue@webmail.hzau.edu.cn

摘要
植物根际微生物群落对植物生长和适逆性至关重要, 本研究对干旱条件下棉花根际真菌群落进行分析, 旨在探明干旱胁迫对棉花根际真菌多样性和群落结构的影响, 为利用有益微生物提高棉花水分利用率提供理论依据。以陆地棉Jin 668 (Gossypium hirsutum cv. Jin668)为试验材料, 采用盆栽控水方式, 对处于开花期的棉花根际土壤(SDP)和未种植棉花土壤(SOPD)进行干旱处理, 正常浇水的棉花根际土壤(SPN)和无棉花土壤(SNPN)为对照。从中采集土壤样品, 提取DNA, 采用Illumina Miseq对真菌ITS1区域进行高通量测序, 研究土壤中真菌多样性。结果共鉴定到970个OTUs, SNPN、SOPD、SPN和SDP样品中真菌OTUs数量分别为481、528、743和752个, 其中288个OTUs为所有组共有。对获得OTUs进行门、纲、目、科和属5个分类水平的划分表明, 棉花根际真菌群落结构主要由子囊菌门(82.70%)和担子菌门(10.15%)组成; 干旱处理使粪壳菌纲(Sordariomycetes)、粪壳菌目(Sordariales)和毛壳菌科(Chaetomiaceae)丰度显著降低, 而散囊菌目(Eurotiales)、发菌科(Trichocomaceae)、曲霉属(Aspergillus)和青霉属(Penicillum)的丰度显著增加。多样性分析结果显示, 与未种棉花的土壤相比, 有棉花的土壤中真菌群落的α多样性显著增加; 同时, SPN和SDP之间的真菌群落结构更相似, 而与SNPN和SOPD间差异较大。研究表明, 棉花根际存在丰富的真菌群落, 干旱对土壤中真菌的丰度和多样性有显著影响。本研究从微生物的角度为提高棉花耐旱性的研究提供新见解。
关键词:
Abstract
Plant rhizosphere microorganisms play important roles in plant growth and the adaptability of plants to adverse environmental stresses. In this study, cotton rhizosphere fungal communities were analyzed under drought conditions, aiming to explore the effects of drought stress on the diversity and community structures of cotton rhizosphere fungi, and to provide a theoretical basis for improving cotton water use efficiency by using beneficial microorganisms. Drought stress was applied to upland cotton (Gossypium hirsutum cv. Jin 668) at flowering stage (SDP), while the soil without plants was also subjected to drought (SOPD). Simultaneously, the plants (SPN) and pots without plants (SNPN) regularly watered were used as controls. The soil samples were collected, the microbial DNA was isolated, and Illumina Miseq was conducted for a high-throughput sequencing of fungi ITS1 regions to study the diversity of the rhizosphere fungal communities. As a result, a total of 970 OTUs were identified, and the numbers of fungal OTUs in the samples of SNPN, SOPD, SPN, and SDP were 481, 528, 743, and 752, respectively, among which 288 OTUs were shared by all samples. The OTUs were classified to different levels of phyla, class, order, family, and genus of fungi. The rhizosphere fungal community of cotton was predominantly consisted of the phyla Ascomycota (82.70%) and Basidiomycota (10.15%). The abundance of Sordariomycetes, Sordariales, and Chaetomiaceae decreased, while the abundance of Eurotiales, Trichocomaceae, Aspergillus, and Penicillum increased significantly under drought stress. Moreover, the diversity of fungal community in the soil with cotton plants significantly higher than that in the soil without cotton plants. Meanwhile, the fungi community structures of SPN and SDP resembling each other and differing greatly from SNPN and SOPD. These results revealed that the cotton rhizosphere had a rich pool of fungal communities, and drought stress had a significant effect on the abundances and diversity of fungi in cotton rhizosphere. This study provided new insights for the researches of improving drought tolerance of cotton in terms of soil microorganisms.
Keywords:
PDF (3679KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
岳丹丹, 韩贝, Abid Ullah, 张献龙, 杨细燕. 干旱条件下棉花根际真菌多样性分析. 作物学报[J], 2021, 47(9): 1806-1815 DOI:10.3724/SP.J.1006.2021.04162
YUE Dan-Dan, HAN Bei, Abid Ullah, ZHANG Xian-Long, YANG Xi-Yan.
干旱是世界粮食安全和植物生产力的主要制约因素之一, 对农作物生长、发育和农艺性状等产生不利影响[1]。棉花是主要经济作物之一, 表现出比其他主要作物如小麦、水稻和玉米对干旱条件更强的耐受性[2]。然而, 干旱等极端环境条件仍然会限制棉花的生长和产量、影响其纤维品质。在长期的进化过程中, 棉花已经形成了一系列形态、生理、生化和分子策略应对干旱胁迫[3]。根系是吸收养分和水分的主要部位, 同时也是分泌排泄物质的重要部位, 植物根系分泌物较多, 营养丰富、微生物多样性丰富[4]。植物影响着其周围及体内微生物的群落结构, 这些微生物又通过其生命活动影响植物的生长发育。微生物组被认为是植物的第2套基因组, 对植物的生长和适逆性至关重要[5]。
研究表明, 特定的根际和内生微生物有助于提高棉花对大丽花轮枝孢菌的抗性[6]。除了保护植物免受生物和非生物胁迫的内在系统外, 在遭受干旱胁迫时, 植物可以与相互作用的微生物(特别是对土壤变化迅速响应的各种土壤微生物群落)建立有益联系, 从而减轻干旱胁迫损伤[7,8], 如来自谷子的耐干旱根际细菌可以在干旱胁迫条件下促进植物生长[9]。同时, 一些体外试验也表明, 根系微生物在植物抵御盐碱、干旱和抗病等非生物胁迫过程中发挥作用[10,11,12]。水稻[13]、玉米[14]、拟南芥[15,16]等多种植物的根系微生物与宿主植物适逆能力相关性的研究也表明有益微生物能提高植物对逆境的适应性。
进一步研究表明, 微生物能合成植物生长调节因子以减轻胁迫对寄主植物的不利影响, 如丛枝菌根真菌可能通过影响宿主植物生长素、细胞分裂素和乙烯等激素分泌, 进而导致植物根系形态发生变化, 增强植物的抗旱性[17]; 水杨酸、茉莉酸、乙烯等植物激素介导的信号通路调控根际促植物生长细菌诱导植物产生对病原菌的免疫反应, 即诱导系统抗病性, 增强植物的抗病性[18]; 基于植物和真菌中赤霉素(gibberellic acids, GA)生物合成途径也被阐明[19]。此外, 微生物还会改变土壤酸碱度、土壤结构、土壤肥力和氧气利用率[20], 如丛枝菌根真菌可通过改善根系形态和改良土壤结构来提高植物的抗旱性[17]。微生物的这些特性可抑制植物病原体并增强植物对高温、盐、干旱和金属毒性的耐受性[21]。同时, 微生物在植物离子转运和营养吸收方面也发挥着重要作用, 如丛枝菌根真菌可以通过改变宿主植株的根系形态和菌丝网络的形成, 以扩大植株对土壤养分吸收范围; 植物释放的有机酸、磷酸酶和质子等根系分泌物可以改变土壤结构和理化性质, 与根际微生物共同作用溶解土壤中难溶的矿质元素(如P和Fe), 提高植株对离子的转运能力、促进其吸收[22]。其次, 微生物还能调控宿主植物的水通道蛋白基因、干旱相
关基因以促进植物在干旱胁迫下的生长和代谢[23], 如丛枝菌根真菌能够特异性诱导苜蓿(Medicago truncatula)水通道蛋白基因的表达, 进而调节苜蓿对硝态氮或铵态氮的吸收[24], 或通过诱导相关磷转运蛋白基因的特异性表达等多种方式, 为植物提供生长和抗逆所需的矿质元素[22,25]。
反过来, 根际微生物群落结构变化也受植物的影响, 如利用高通量测序技术测定的花铃期土壤中细菌群落结构结果表明, 黄萎病抗病品种在花铃期的细菌群落结构优于感病品种[12]。此外, 根际微生物的群落结构也受其他多种因素影响。在不同的发育阶段, 棉花根际细菌的群落结构变化很大[26]; 土壤微生物群落对长期连作棉花也有相似的反应, 即在相同的生境中, 微生物群落表现出相似性[27]; 相对无植物种植的土壤, 即使在干旱胁迫下棉花根际也有着丰富的细菌群落, 这可能有助于提高植物的耐旱性[28]。
植物-微生物互作为研究植物适逆性提供了一个新视角, 对有益微生物的使用将有助于农业生产系统走向一个更具生产力的时代。目前, 高通量测序方法为根际微生物群落的研究提供了新的思路和方法。本研究利用高通量测序技术, 通过对正常和干旱条件下棉花根际真菌群落特征进行分析, 研究干旱对棉田土壤中真菌多样性和群落结构的影响, 确定最适合棉花抗旱研究的真菌种类, 有助于挖掘棉田中有益微生物, 同时对提高棉花耐旱性的研究具有重要的理论意义。
1 材料与方法
1.1 植物材料、干旱处理及样品采集
土壤的收集、材料的种植及干旱处理参照文献[28], 从长期连作棉花试验田采集土壤, 将土壤在阳光下干燥1个月, 然后用2 mm的筛子过筛, 称取2.5 kg加入每个土钵中备用。棉花种子(陆地棉Jin 668)用0.1%氯化汞消毒, 在棉花无菌苗培养基中萌发, 炼苗后转入上述土钵中, 每个土钵种1棵植株, 平行设置不种棉花的土钵土壤, 所有的土壤均用蒸馏水灌溉。植株在温室条件下[(28±2)℃, 约16 h/8 h光/暗光周期]生长, 在进入六叶期, 对种植棉花的土钵和不种棉花的土钵进行干旱处理(不浇水), 以正常浇水的有棉花的土钵和无棉花的土钵为对照, 干旱处理2周后, 干旱处理的棉花植株发生萎焉, 此时收集各组土壤样品, 有棉花种植的土壤微生物样品由根表土壤微生物和根围土壤微生物组成。棉花根表土壤微生物的收集方法如下: 小心收集干旱和对照棉花的根系, 然后放置在含有25 mL磷酸盐缓冲液(每升6.33 g NaH2PO4·H2O, 16.5 g Na2HPO4·7H2O, 200 mL Silwet L-77)的50 mL无菌试管中, 高速离心去除根表土壤; 将上层溶液通过100 mm的尼龙膜过滤器过滤以去除残渣, 然后在5000 × g转速下离心15 min, 收集下层含有微生物的沉淀, 迅速在液氮中冷冻, 然后-80℃储存。根围土壤样品的收集: 在棉株周围(无棉株)土壤中6个方向均匀取样, 15个土钵为一个重复进行混合, 然后收混集10 g土壤样品放入无菌离心管中。共获得4种土壤微生物样品, 分别为干旱处理的棉花根际土壤样品(drought treated plant rhizosphere soil, SDP)、正常浇水棉花根际土壤样品(normally watered plant rhizosphere soil, SPN)、未种棉花干旱处理的土壤(drought treated soil without plants, SOPD)和正常浇水的土壤(normally watered soil without plants, SNPN)样品。每次处理进行3次生物重复, 共收集12个样品。
1.2 DNA提取、真菌ITS1区扩增、Illumina文库的制备和高通量测序
由上海派森诺生物科技股份有限公司(中国上海)完成样品微生物DNA的提取、Illumina文库的制备和高通量测序。通过引物ITS5F (5°-GGAAGTAAAAGTCGTAA CAAGG-3°)和ITS2R (5°-GCTGCGTTCTTCATCGATGC-3°)扩增真菌ITS1区, 采用Illumina MiSeq测序平台进行双末端(Paired-end)测序。1.3 原始数据处理、操作分类单位(OTU)划分及多样性分析
利用QIIME 1.8.0软件对测序数据进行识别、拼接, 并去除低质量、非特异扩增序列及嵌合体序列[29]。在相似性97%的水平上使用UCLUST对获得的序列进行操作分类单元(operational taxonomic unit, OTU)聚类分析, 并筛选出OTUs的代表性序列[30]; 基于UNITE数据库(Release 5.0,β多样性分析用于检验不同样本之间群落结构的相似性, 主要通过主成分分析(principal component analysis, PCA)和多维尺度分析(multidimensional scaling, MDS)进行。
1.4 统计分析
观察评价原始样本组间差异大小及其统计学显著性是通过基于排列的PERMANOVA分析来评估。Adonis/ PERMANOVA多元方差分析使用QIIME软件进行, 并进行999次置换检测组间差异是否具有统计学显著性。UniFrac距离组间或组内差异比较分析是根据样本分组, 分别对加权的UniFrac距离矩阵的组内和组间距离均值进行t检验, 并通过1000次蒙特卡罗置换检验判断统计显著性。2 结果与分析
2.1 高通量测序结果分析
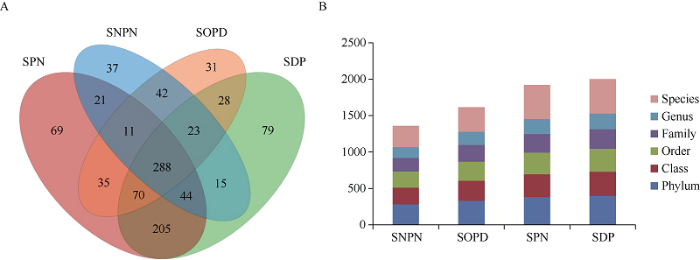
为了探索正常和干旱条件下棉花根际真菌群落, 对正常和干旱条件下的棉花根际土壤和无棉花种植的土壤共12个样品进行菌群DNA抽提, 利用Illumina高通量测序技术对真菌ITS1区进行测序, 共获得568,592条Clean tags, 并进行OTUs的划分。基于≥97%的相似度水平, 通过聚类共获得970个用于物种分类的OTUs, 其中SNPN、SOPD、SPN和SDP的OTUs总数分别为481、528、743和752个, 其中288个OTUs为所有组共有(图1-A)。在OTU鉴定后, 对每个样品的OTUs划分为门、纲、目、科、属和种5个分类水平, 结果表明, 相对无棉花种植的土壤, 棉花根际土壤真菌群落更丰富; 另外, 相对于对照, 干旱处理也增加了土壤中微生物的多样性(图1-B)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1微生物群落的OTU分类
A: 样本OTUs数量的Venn图; B:不同分类级别下样本OTUs数。SNPN: 正常浇水的土壤样品; SOPD: 未种棉花干旱处理的土壤样品; SPN: 正常浇水棉花根际土壤样品; SDP: 干旱处理的棉花根际土壤样品。
Fig. 1OTU classification of microbial communities
A: Venn diagram of sample OTUs quantity; B: the number of OTUs of sample at different classification level. SNPN: normally watered soil without plants; SOPD: drought treated soil without plants; SPN: normally watered plant rhizosphere soil; SDP: drought treated plant rhizosphere soil.
2.2 α多样性分析
通过进行α多样性分析检查单个微生物生态系统中物种丰富度和均匀度。真菌群落稀疏曲线结果显示, 序列量到5000时各样品稀释曲线均基本趋于平缓, 表明取样基本合理, 真实环境中真菌群落结构的置信度较高, 能够比较真实地反映土壤样本的真菌群落(图2-A)。同时在抽取相同序列的条件下, SOP和SPN相对SOPN和SNPN样本的OUT数多, 说明SOP和SPN样本中物种丰富度更高。此外, 物种累积曲线也显示了样本量足以反映种群的丰富度(图2-B)。等级丰度曲线也表明, SPN和SDP具有更高的均匀度和丰富度, 而SNPN和SOPD在OTUs上的丰度差异更大且均匀度较低(图2-C)。图2
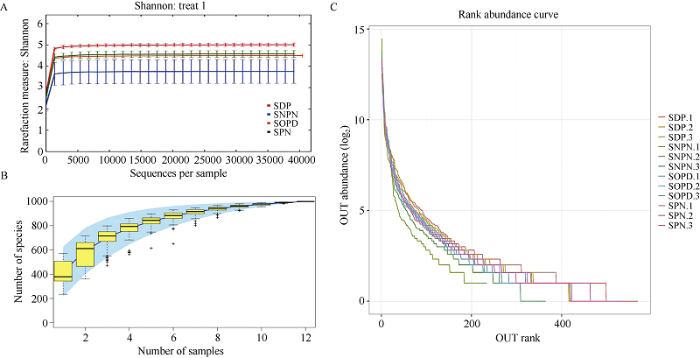 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2正常和干旱下土壤真菌α多样性分析
A: 稀疏曲线; B: 物种累积曲线; C: 丰度等级曲线。缩写同
Fig. 2Alpha diversity analysis of soil fungi under normal and drought conditions
A: sparse curves, B: species cumulative curve; C: rank abundance curve. Abbreviations are the same as those given in
许多指数可以反映微生物群落的α多样性。Shannon指数反映群落中物种的变化度, Shannon指数高代表群落中物种丰富度高且分布均匀, 常用的测量指标还包括反映群落丰富度的Chao 1指数和ACE指数, 以及反映群落均匀度的Simpson指数。分别计算每个样本的α多样性指数, 并在表1中列出。结果显示, SNP和SDP的α多样性指数与对照相比均显著地提高, 表明SPN和SDP具有较高的物种丰富度和均匀度。
Table 1
表1
表1微生物多样性指数
Table 1
| 样品 Sample | Chao1指数 Chao1 index | ACE指数 ACE index | 辛普森指数 Simpson index | 香农指数 Shannon index |
|---|---|---|---|---|
| SNPN | 304.33±15.04 b | 316.50±19.09 b | 0.79±0.11 b | 4.14±0.29 b |
| SOPD | 334.50±12.02 b | 334.50±12.02 b | 0.89±0.00 b | 4.50±0.09 b |
| SPN | 544.40±14.91 a | 567.03±15.54 a | 0.90±0.02 a | 4.58±0.19 a |
| SDP | 525.31±17.30 a | 527.92±14.83 a | 0.92±0.01 a | 5.01±0.10 a |
新窗口打开|下载CSV
2.3 分类学分析
基于OTU分类结果, 获得了门、纲、目、科、属和种分类水平下每个样品的特有组成。不同样品在每一个分类级别下的微生物类群都有差异, 总体上SDP在纲、目、属和种水平上比其他样本具有更高的微生物类群数量(表2)。Table 2
表2
表2各分类水平的微生物类群数统计表
Table 2
| 样品 Sample | 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species |
|---|---|---|---|---|---|---|
| SNPN | 9.00±0.00 a | 15.00±1.00 b | 39.33±2.89 b | 62.50±3.54 a | 88.50±2.12 b | 158.00±2.83 d |
| SOPD | 8.67±1.15 a | 15.67±3.06 ab | 43.00±5.00 ab | 67.00±2.00 a | 95.67±4.04 a | 168.00±1.41 c |
| SPN | 9.00±0.00 a | 18.67±1.53 ab | 45.00±4.58 a | 68.67±2.08 a | 103.50±2.83 a | 182.00±1.41 b |
| SDP | 9.00±0.00 a | 20.00±1.00 a | 51.00±4.58 a | 70.00±0.00 a | 109.00±2.83 a | 194.00±1.41 a |
新窗口打开|下载CSV
将OTU的代表序列与微生物参考数据库进行比对可得到每个OTU对应的物种分类信息, 进而获得了5个分类层次下(门、纲、目、科和属)每个样品的组成和丰度分布。分析棉花土壤样品真菌多样性发现, 门分类水平上, 大多数真菌是子囊菌门(Ascomycota)、担子菌门(Basidiomycota)、类原生动物门(Rozellomycota)、接合菌门(Zygomycota)、纤毛亚门(Ciliophora)、壶菌门(Chytridiomycota)、球囊菌门(Glomeromycota), 其中子囊菌门(Ascomycota)为第一优势门。与无棉花种植的土壤相比, 棉花根际土壤中壶菌门(Chytridiomycota)和球囊菌门(Glomeromycota)丰度显著降低(图3-A)。
图3
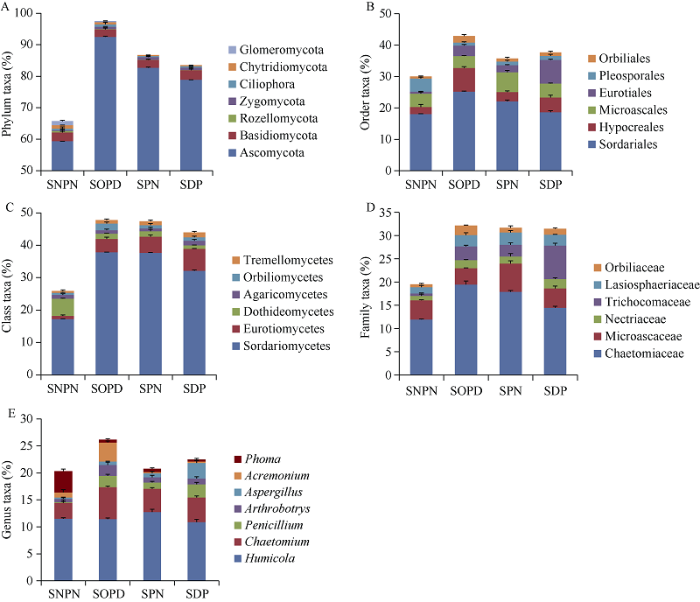 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3分类单元的分布和丰度图
A~E表示分类单元在门、纲、目、科和属水平上的百分比。缩写同
Fig. 3Distribution and abundance of taxa
A-E represent the percentage of taxa at phylum, class, order, family, and genus level, respectively. Abbreviations are the same as those given in
在纲分类水平上, 大多数真菌是粪壳菌纲(Sordariomycetes)、散囊菌纲(Eurotiomycetes)、座囊菌纲(Dothideomycetes)、伞菌纲(Agaricomycetes)、圆盘菌纲(Orbiliomycetes)、银耳纲(Tremellomycetes)。与无棉花种植的土壤相比, 散囊菌纲(Eurotiomycetes)在棉花根际土壤中的丰度显著增加, 而粪壳菌纲(Sordariomycetes)和座囊菌纲(Dothideomycetes)的丰度显著降低。此外, 与正常浇水处理的棉花根际土壤相比, 粪壳菌纲(Sordariomycetes)在干旱处理的棉花根际土壤中的丰度显著降低(图3-B)。
在目分类水平上, 大多数真菌是粪壳菌目(Sordariales)、肉座菌目(Hypocreales)、小囊菌目(Microascales)、散囊菌目(Eurotiales)、格孢腔菌目(Pleosporales)、圆盘菌目(Orbiliales)。与无棉花种植的土壤相比, 棉花根际土壤中的散囊菌目(Eurotiales)丰度显著增加, 而肉座菌目(Hypocreales)和格孢腔菌目(Pleosporales)的丰度显著降低。此外, 与正常浇水处理的棉花根际土壤相比, 散囊菌目(Eurotiales)在干旱处理的棉花根际中丰度显著增加, 而粪壳菌目(Sordariales)降低(图3-C)。
毛壳菌科(Chaetomiaceae)、小囊菌科(Microascaceae)、丛赤壳科(Nectriaceae)、发菌科(Trichocomaceae)、毛球壳科(Lasiosphaeriaceae)、圆盘菌科(Orbiliaceae)是较为丰富的科。相对无棉花种植的土壤, 棉花根际土壤中发菌科(Trichocomaceae)和毛球壳科(Lasiosphaeriaceae)的丰度显著增加, 而毛壳菌科(Chaetomiaceae)的丰度显著降低。此外, 在干旱处理的棉花根际中发菌科(Trichocomaceae)的丰度显著增加, 而毛壳菌科(Chaetomiaceae)降低(图3-D)。
在属分类水平上, 腐质霉属(Humicola)是棉花根际土壤真菌的优势菌属, 其在SPN土壤中相对丰度最高, 而在无棉花种植的土壤中相对丰度最低。此外, 毛壳菌属(Chaetomium)、青霉属(Penicillum)、节丛孢属(Arthrobotrys)、曲霉属(Aspergillus)、顶孢霉属(Acremonium)和茎点霉属(Phoma)也是不同处理下土壤中的优势菌群。与无棉花种植的土壤相比, 曲霉属(Aspergillus)和青霉属(Penicillum)在棉花根际土壤中占优势, 而顶孢霉属(Acremonium)和茎点霉属(Phoma)的丰度降低。干旱处理增加了曲霉属(Aspergillus)和青霉属(Penicillum)的丰度(图3-E)。
系统发育树分析表明, 子囊菌门(Ascomycota)和担子菌门(Basidiomycota)是棉花根际土壤中最丰富的门。此外, 最丰富的纲是粪壳菌纲(Sordariomycetes)和伞菌纲(Agaricomycetes) (图4-A)。根据分类群的丰度分布或样品之间的相似程度, 对前50个属在不同样品中进行聚类分析, 与无棉花种植的土壤相比, 绿僵菌属(Metarhizium)、曲霉属(Aspergillus)、斜盖伞属(Clitopilus)、青霉属(Penicillium)、柄孢壳属(Zopfiella)、节担菌属(Wallemia)、根霉属(Rhizopus)、足放线病菌属(Scedosporium)和蛇口壳属(Ophiostoma)在棉花的根际土壤中相对丰富(图4-B)。
图4
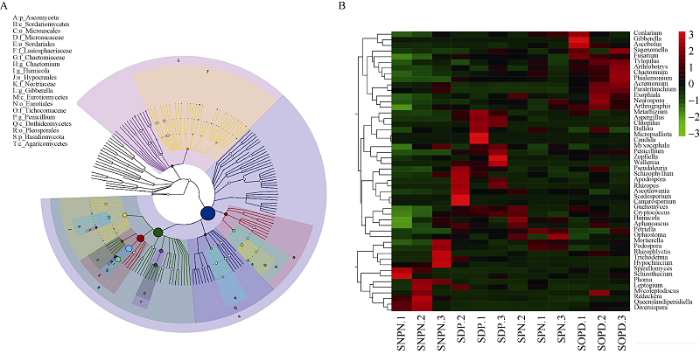 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4系统进化树和热图的分类分析
A: 显示样本群体中从门到属水平的所有分类群的层次关系的分类层次树; B: 综合群体组成的热水平图和聚类分析。缩写同
Fig. 4Taxonomic analysis of phylogenetic tree and heat map
A: the classification hierarchy tree shows the hierarchical relationships of all taxa from the phylum to the genus level in the sample population; B: combined heat level map of the community composition with cluster analysis. Abbreviations are the same as those given in
2.4 β多样性分析
为了观察不同样本之间真菌群落的相似性, 通过主成分分析(PCA)和多维尺度分析对12个样本进行β多样性分析。PCA分析发现, 第1主成分(PCA1)和第2主成分(PCA2)分别可以解释所有变量的74.15%和13.32%, 2个主成分方差累积贡献率达到87.47%, 说明其能够表征微生物群落组成的特征, 进一步分析表明, SPN和SDP棉花根际样品的微生物群落结构相对相似(图5-A)。经加权UniFrac PCoA分析显示, 与没有种植棉花的土壤样品(SNPN和SOPD)相比, SPN和SDP样品的真菌群落结构具有很高的相似性, 并且不同(图5-B)。为更全面地描述组内和组间的菌群结构差异大小, 计算各组之间和组内加权的UniFrac距离均值。结果显示, SPN-SNPN和SDP-SNPN之间有很大的UniFrac距离, 然而, SDP-SPN之间UniFrac距离较小, 特别是在SPN、SDP和SOPD组内, UniFrac距离非常小(图5-C)。图5
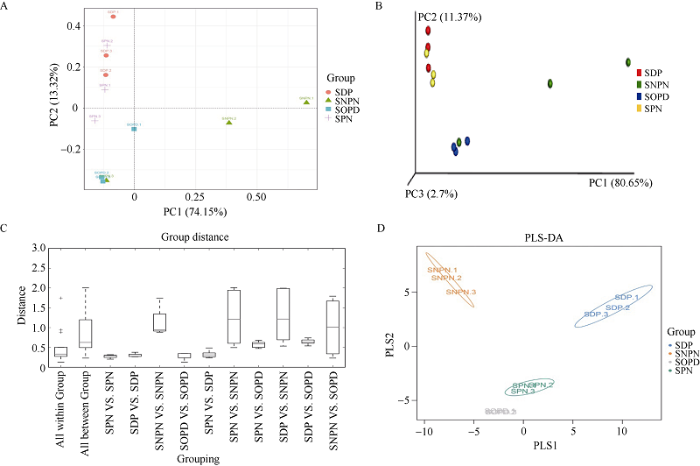 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5β多样性分析
A: 主成分分析的二维排序; B: 加权UniFrac的PCoA分析; C: 加权UniFrac距离的多组方框图; D: PLS-判别式分析。缩写同
Fig. 5Beta diversity analysis
A: two-dimensional ranking of the PCA analysis; B: weighted UniFrac PCoA analysis; C: multiple sets of box plots for the weighted UniFrac distance; D: PLS-discriminant analysis. Abbreviations are the same as those given in
同时我们也利用偏最小二乘法判别分析(partial least squares discrimination analysis, PLS-DA)对基于物种丰度矩阵和样本分组数据构建的高通量测序产生的大量微生物群落数据进行判别分析显示, SPN和SOPD彼此之间更相似(图5-D)。此外, 对原始样本组间的差异还进行了方差分析, 加权UniFrac的R统计值为0.7253。此外, 加权后的UniFrac的P值和排列数分别为0.001和999。
3 讨论
全球环境变化对植物生长产生不利影响, 微生物组作为植物的第2套基因组, 对植物生长及适逆性非常重要, 因此微生物群落在增强植物生长方面的潜在功能使得阐明植物微生物群落对环境变化的响应变得非常重要。微生物群落不是一成不变的, 受到环境以及微生物间和植物与微生物间交流的调节[33]。根系分泌物吸引附近土壤环境中的微生物, 改变的根系分泌物扩散到土壤后, 影响了作物根际的土壤环境, 从而致使作物根际微生物数量及种类发生特定变化。例如, 不同植物之间微生物群落组成有显著差异[20], 环境条件如干旱、盐度、酸碱度、土壤结构和肥力也影响微生物群落[16,34]。研究表明, 有机肥处理后土壤中细菌和放线菌的群落多样性与Shannon指数显著升高, 且氮肥处理后真菌多样性水平升高[35]; 干旱胁迫的发生也能够引起棉花根际土壤中细菌的富集且群落多样性显著增加[28]。在本研究中, 通过微生物基因的高通量测序来检测干旱和正常条件下棉花根际及无棉花种植的连作棉田土壤中的真菌群落。研究结果表明, 不同处理土壤中真菌群落的主要成分存在差异, 其中分类学上不同种类真菌的相对丰度发生变化, 棉花根际真菌群落结构主要由子囊菌门(Ascomycota)、担子菌门(Basidiomycota)、类原生动物门(Rozellomycota)、接合菌门(Zygomycota)、纤毛亚门(Ciliophora)、壶菌门(Chytridiomycota)、球囊菌门(Glomeromycota)组成, 其中子囊菌门和担子菌门是植物根际土壤中最丰富的真菌门, 这一结果与前人研究一致[36]。子囊菌门是土壤微生物群落中最丰富的真菌门, 具有降解有机物质的作用, 在有机质含量高的土壤中含量丰富[26,37]。
聚类分析结果表明, 在正常和干旱条件下, 与无棉花种植的土壤相比, 棉花根际土壤中绿僵菌属(Metarhizium)、曲霉属(Aspergillus)、斜盖伞属(Clitopilus)、青霉属(Penicillium)、柄孢壳属(Zopfiella)、节担菌属(Wallemia)、根霉属(Rhizopus)等相对丰富。其中, 曲霉属(Aspergillus)和青霉属(Penicillium)在干旱胁迫下棉花根际土壤中丰度较高。这与前人发现凤仙花的根际真菌群落中青霉属, 曲霉属等优势种在旱季出现的频度最高这一研究结果一致[38]。曲霉属是盐场高浓度盐水中本土真菌群落的一部分[39], 具有更高的胁迫耐受性, 其中曲霉属的构巢曲霉(Aspergillus nidulans)具有耐旱性[40]。此外, 研究发现曲霉属和青霉属有较强的渗透压耐受性, 耐受性最强的是菌种多属于曲霉属[41]。因此, 这些对干旱敏感的微生物, 尤其是在干旱条件下植物根际富集的微生物可能有助于植物抵抗干旱和其他非生物胁迫。
近年来研究已报道, 植物在遭受干旱胁迫时, 根际促植物生长细菌作用的植物能更快速产生高量的ABA, 更快地调控基因表达和代谢, 积累脯氨酸和甜菜碱等渗透调节物质, 或增加抗氧化酶和抗氧化物, 提高抗氧化酶活性, 增强植物耐旱的能力[25]。同时, 激素作为植物和微生物之间的中介, 影响植物微生物群落结构的功能[33], 在植物遭受非生物胁迫时也发挥着积极的作用[42]。如根际促植物生长细菌能够合成刺激植物细胞生长和分裂的植物激素, 从而促使植物抵御环境胁迫[18]。因此, 我们推测曲霉属(Aspergillus)和青霉属(Penicillium)真菌可能通过刺激植物合成更多的ABA或植物激素的方式, 参与调节植物在抗旱的信号通路, 以提高植物的抗旱性。此外, 它们对植物抗旱性具体作用机制有待进一步的研究。
本研究还发现, 在干旱和正常条件下, 棉花的种植与否对根际真菌群落结构的α多样性有不同的影响。棉花的种植提升了土壤真菌的物种多样性和物种丰富度, SPN和SPD样品具有较高的均匀度和丰富度, 而SNPN和SOPD的OTUs丰度差异较大且均匀度较低。同时, 在干旱胁迫下, SDP有较高的OTUs。这些结果均表明, 植物根系物质的分泌改变了微生物生态环境, 吸引着附近土壤环境中的微生物富集, 使得根际微生物的物种更多样, 丰富度更高。这些结果与前人发现棉花根际具有更丰富的细菌群落的结果一致[28]; 另一方面, 干旱胁迫可能通过调节根际分泌物的释放, 刺激了土壤中一些微生物的生长, 在变化的环境下维持植物适应性[43]。此外, β多样性结果显示, SPN和SDP之间的微生物群落结构更相似, 与SNPN和SOPD间差异较大, 同样, SNPN和SOPD彼此之间更相似。结果表明棉花的种植改变了根际土壤微生物群落结构。棉花根际真菌群落结构的变化可能主要受到棉花根与土壤相互作用的影响 [28,33,43], 植物根系改变土壤的物理和化学性质, 从而致使根际微生物数量及种类发生特定变化[44]。无棉花种植的土壤(SNPN和SOPD)和根际土壤(SPN和SDP)微生物多样性的差异分析表明, 微生物附着在根上, 可能随后促进植物的生长发育、增强耐旱性。此外, 在这项研究中, 鉴定出的许多真菌与棉花植物之间的具体关系是未知的, 因此, 需要进行更多的研究, 尤其是侧重于天然存在的根际真菌, 这些真菌是生物防治剂的潜在来源。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

The growth and yield of many crops, including cotton, are affected by water deficit. Cotton has evolved drought specific as well as general morpho-physiological, biochemical and molecular responses to drought stress, which are discussed in this review. The key physiological responses against drought stress in cotton, including stomata closing, root development, cellular adaptations, photosynthesis, abscisic acid (ABA) and jasmonic acid (JA) production and reactive oxygen species (ROS) scavenging, have been identified by researchers. Drought stress induces the expression of stress-related transcription factors and genes, such as ROS scavenging, ABA or mitogen-activated protein kinases (MAPK) signalling genes, which activate various drought-related pathways to induce tolerance in the plant. It is crucial to elucidate and induce drought-tolerant traits via quantitative trait loci (QTL) analysis, transgenic approaches and exogenous application of substances. The current review article highlights the natural as well as engineered drought tolerance strategies in cotton. © 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]

wilt caused by is a common soil-borne disease worldwide, affecting many economically important crop species. Soil microbes can influence plant disease development. We investigated rhizosphere and endosphere microbiomes in relation to cotton cultivars with differential susceptibility to wilt. Soil samples from nine cotton cultivars were assessed for the density of microsclerotia; plants were assessed for disease development. We used amplicon sequencing to profile both bacterial and fungal communities. Unlike wilt severity, wilt inoculum density did not differ significantly among resistant and susceptible cultivars. Overall, there were no significant association of alpha diversity indices with wilt susceptibility. In contrast, there were clear differences in the overall rhizosphere and endosphere microbial communities, particularly bacteria, between resistant and susceptible cultivars. Many rhizosphere and endosphere microbial groups differed in their relative abundance between resistant and susceptible cultivars. These operational taxonomic units included several well-known taxonomy groups containing beneficial microbes, such as Bacillales, Pseudomonadales, Rhizobiales, and, which were higher in their relative abundance in resistant cultivars. Greenhouse studies with sterilized soil supported that beneficial microbes in the rhizosphere contribute to reduced wilt development. These findings suggested that specific rhizosphere and endosphere microbes may contribute to cotton resistance to. Copyright © 2019 Wei, Zhao, Xu, Feng, Shi, Deakin, Feng and Zhu.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOIPMID [本文引用: 2]

Beneficial microbes in the microbiome of plant roots improve plant health. Induced systemic resistance (ISR) emerged as an important mechanism by which selected plant growth-promoting bacteria and fungi in the rhizosphere prime the whole plant body for enhanced defense against a broad range of pathogens and insect herbivores. A wide variety of root-associated mutualists, including Pseudomonas, Bacillus, Trichoderma, and mycorrhiza species sensitize the plant immune system for enhanced defense without directly activating costly defenses. This review focuses on molecular processes at the interface between plant roots and ISR-eliciting mutualists, and on the progress in our understanding of ISR signaling and systemic defense priming. The central role of the root-specific transcription factor MYB72 in the onset of ISR and the role of phytohormones and defense regulatory proteins in the expression of ISR in aboveground plant parts are highlighted. Finally, the ecological function of ISR-inducing microbes in the root microbiome is discussed.
DOIURL [本文引用: 1]
DOIPMID [本文引用: 2]

After a century of incremental research, technological advances, coupled with a need for sustainable crop yield increases, have reinvigorated the study of beneficial plant-microbe interactions with attention focused on how microbiomes alter plant phenotypes. We review recent advances in plant microbiome research, and describe potential applications for increasing crop productivity. The phylogenetic diversity of plant microbiomes is increasingly well characterized, and their functional diversity is becoming more accessible. Large culture collections are available for controlled experimentation, with more to come. Genetic resources are being brought to bear on questions of microbiome function. We expect that microbial amendments of varying complexities will expose rules governing beneficial plant-microbe interactions contributing to plant growth promotion and disease resistance, enabling more sustainable agriculture. Copyright © 2017 The Authors. Published by Elsevier Ltd.. All rights reserved.
DOIPMID [本文引用: 1]

Land plants associate with a root microbiota distinct from the complex microbial community present in surrounding soil. The microbiota colonizing the rhizosphere (immediately surrounding the root) and the endophytic compartment (within the root) contribute to plant growth, productivity, carbon sequestration and phytoremediation. Colonization of the root occurs despite a sophisticated plant immune system, suggesting finely tuned discrimination of mutualists and commensals from pathogens. Genetic principles governing the derivation of host-specific endophyte communities from soil communities are poorly understood. Here we report the pyrosequencing of the bacterial 16S ribosomal RNA gene of more than 600 Arabidopsis thaliana plants to test the hypotheses that the root rhizosphere and endophytic compartment microbiota of plants grown under controlled conditions in natural soils are sufficiently dependent on the host to remain consistent across different soil types and developmental stages, and sufficiently dependent on host genotype to vary between inbred Arabidopsis accessions. We describe different bacterial communities in two geochemically distinct bulk soils and in rhizosphere and endophytic compartments prepared from roots grown in these soils. The communities in each compartment are strongly influenced by soil type. Endophytic compartments from both soils feature overlapping, low-complexity communities that are markedly enriched in Actinobacteria and specific families from other phyla, notably Proteobacteria. Some bacteria vary quantitatively between plants of different developmental stage and genotype. Our rigorous definition of an endophytic compartment microbiome should facilitate controlled dissection of plant-microbe interactions derived from complex soil communities.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
DOIURL [本文引用: 2]
DOIURL [本文引用: 1]
DOIPMID [本文引用: 5]

Climate change contributes to drought stress and subsequently affects crop growth, development, and yield. The microbial community, such as fungi and bacteria in the rhizosphere, is of special importance to plant productivity. In this study, soil collected from a cotton research field was used to grow cotton plants (Gossypium hirsutum cv. Jin668) under controlled environment conditions. Drought stress was applied at flowering stage, while control plants were regularly watered. At the same time, the soil without plants was also subjected to drought, while control pots were regularly watered. The soil was collected in sterilized tubes and microbial DNA was isolated and high-throughput sequencing of 16S rRNA genes was carried out. The alpha diversity of bacteria community significantly increased in the soil with cotton plants compared to the soil without cotton plants. Taxonomic analysis revealed that the bacterial community structure of the cotton rhizosphere predominantly consisted of the phyla Proteobacteria (31.7%), Actinobacteria (29.6%), Gemmatimonadetes (9.8%), Chloroflexi (9%), Cyanobacteria (5.6%), and Acidobacteria. In the drought-treated rhizosphere, Chloroflexi and Gemmatimonadetes were the dominant phyla. This study reveals that the cotton rhizosphere has a rich pool of bacterial communities even under drought stress, and which may improve drought tolerance in plants. These data will underpin future improvement of drought tolerance of cotton via the soil microbial community.
DOIPMID [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 3]
DOIPMID [本文引用: 1]

Plants have incredible developmental plasticity, enabling them to respond to a wide range of environmental conditions. Among these conditions is the presence of plant growth-promoting rhizobacteria (PGPR) in the soil. Recent studies show that PGPR affect Arabidopsis thaliana root growth and development by modulating cell division and differentiation in the primary root and influencing lateral root development. These effects lead to dramatic changes in root system architecture that significantly impact aboveground plant growth. Thus, PGPR may promote shoot growth via their effect on root developmental programs. This review focuses on contextualizing root developmental changes elicited by PGPR in light of our understanding of plant-microbe interactions and root developmental biology. Copyright © 2016 Elsevier Ltd. All rights reserved.
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOIURL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
PMID [本文引用: 1]

Six different species of the known teleomorphic food-borne xerophilic genus Eurotium were repeatedly isolated in a mycodiversity study of hypersaline waters. At salinities above 17% NaCl, E. amstelodami was detected most consistently, followed by E. repens and E. herbariorum, while E. rubrum, E. chevalieri and a potentially new species, "Eurotium halotolerans", were detected only occasionally at lower salinities. The qualitative secondary metabolite profiles produced by Eurotium spp. from salterns were not different from those of Eurotium spp. from foods and other habitats. Spatiotemporal frequency of occurrence and in vitro determined adaptive ability of propagules to survive prolonged exposure to hypersaline conditions indicate that E. amstelodami, E. herbariorum, and E. repens contribute to the indigenous fungal community in hypersaline water environments, while E. rubrum, E. chevalieri and "E. halotolerans" are only temporal inhabitants of brine at lower salinities.
PMID [本文引用: 1]

Aspergillus nidulans was shown to be xerotolerant, with optimal radial growth on basal medium amended with 0.5 M NaCl (osmotic potential [psi s] of medium, -3 MPa), 50% optimal growth on medium amended with 1.6 M NaCl (psi s of medium, -8.7 MPa), and little growth on medium amended with 3.4 M NaCl (psi s of medium, -21 MPa). The intracellular content of soluble carbohydrates and of selected cations was measured after growth on basal medium, on this medium osmotically amended with NaCl, KCl, glucose, or glycerol, and also after hyperosmotic and hypoosmotic transfer. The results implicate glycerol and erythritol as the major osmoregulatory solutes. They both accumulated during growth on osmotically amended media, as well as after hyperosmotic transfer, except on glycerol-amended media, in which erythritol did not accumulate. Furthermore, they both decreased in amount after hypoosmotic transfer. With the exception of glycerol, the extracellular osmotic solute did not accumulate intracellularly when mycelium was grown in osmotically amended media, but it accumulated after hyperosmotic transfer. It was concluded that the extracellular solute usually plays only a transient role in osmotic adaptation. The intracellular content of soluble carbohydrates and cations measured could reasonably account for the intracellular osmotic potential of mycelium growing on osmotically amended media.
DOIURL [本文引用: 1]
DOIURL [本文引用: 1]
DOIURL [本文引用: 2]
[本文引用: 1]
