 ,1,2, 娄鸿耀1, 陈朝燕1, 肖菁2, 徐麟2, 倪中福1, 刘杰
,1,2, 娄鸿耀1, 陈朝燕1, 肖菁2, 徐麟2, 倪中福1, 刘杰 ,1,*
,1,*Genetic diversity assessment of winter wheat landraces and cultivars in Xinjiang via SNP array analysis
MA Yan-Ming ,1,2, LOU Hong-Yao1, CHEN Zhao-Yan1, XIAO Jing2, XU Lin2, NI Zhong-Fu1, LIU Jie
,1,2, LOU Hong-Yao1, CHEN Zhao-Yan1, XIAO Jing2, XU Lin2, NI Zhong-Fu1, LIU Jie ,1,*
,1,*通讯作者:
收稿日期:2019-12-27接受日期:2020-03-24网络出版日期:2020-04-28
| 基金资助: |
Received:2019-12-27Accepted:2020-03-24Online:2020-04-28
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (458KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
马艳明, 娄鸿耀, 陈朝燕, 肖菁, 徐麟, 倪中福, 刘杰. 新疆冬小麦地方品种与育成品种基于SNP芯片的遗传多样性分析[J]. 作物学报, 2020, 46(10): 1539-1556. doi:10.3724/SP.J.1006.2020.91077
MA Yan-Ming, LOU Hong-Yao, CHEN Zhao-Yan, XIAO Jing, XU Lin, NI Zhong-Fu, LIU Jie.
小麦(Triticum aestivum L.)是我国三大主粮作物之一, 种植面积广、总产量高[1]。新疆是我国西北重要的小麦优势产区和消费区, 冬小麦播种面积占全区小麦总面积的70%以上[2,3]。新中国成立以来, 新疆南北疆的冬小麦种植区都经历了“地方品种—引进品种—自育品种”的更替。事实证明, 小麦品种改良在增产中取得了巨大成效。然而, 随着小麦品种产量潜力不断提高, 新育成品种也出现了品种类型相似和抗病、抗逆性较差的问题, 在产量和适应性上没有大的突破[4,5]。这可能与育种过程中长期对产量、抗性、品质等性状的连续定向选择, 造成育成品种遗传基础日益狭窄、遗传多样性下降有关。遗传变异是遗传育种的基础, 认识和了解遗传变异将对育种过程中杂交组合的合理选配以及后代选择提供指导。因此, 非常有必要结合利用最新的分子标记技术手段, 对新疆的冬小麦地方品种和育成品种(系)及其育种亲本材料的遗传基础和亲缘关系全面深入了解, 为亲本选配、分子标记辅助选择和小麦育种实践提供应用与指导。
单核苷酸多态性(single nucleotide polymorphism, SNP)是美国****Lander于1996年提出的第3代DNA遗传标记[6], 因与芯片技术和基因克隆相结合, 具有数量多、易于自动化检测分型和通量高等优点, 在小麦遗传图谱构建、基因定位、遗传多样性分析、农艺性状关联分析、分子标记辅助育种等方面发挥着重要作用。随着高通量测序技术的快速发展, 先后开发出小麦9K、90K、660K、55K等小麦SNP芯片, 其中小麦55K SNP芯片是由中国农科院作物科学研究所小麦基因组与基因资源研究团队在小麦660K SNP芯片的基础上开发的, 经过了数千份国内小麦品种材料的检验, 相较于9K、90K和660K芯片, 其有效标记的比例更高, 具有更高的利用价值[7,8]。目前利用小麦55K SNP芯片仅见于构建高密度遗传连锁图[6,7]以及关联分析方面[9,10], 尚未见有关利用小麦55K SNP芯片开展小麦种质遗传多样性研究的报道。
由于小麦品种具有很强的区域适应性, 在不同种植区具有明显的地区特征和习性, 各地区育种目标也不完全相同, 针对特定区域的小麦品种材料及育种实践开展有关研究非常必要。本研究利用小麦55K SNP标记技术揭示134个新疆本土冬小麦地方品种和54个新疆冬小麦育成品种的遗传多样性和亲缘关系, 并探讨新疆冬小麦品种在改良过程中基因多样性的变化趋势, 全面阐述新疆冬小麦种质的遗传特性, 为小麦育种实践提供指导和参考。
1 材料与方法
1.1 试验材料及DNA提取
选用188个新疆冬小麦品种资源, 其中地方品种134个, 育成品种54个, 均为新疆农业科学院农作物品种资源研究所收集保存的材料, 试验采用2015年10月种植, 2016年7月收获的自花授粉种子。全部试验材料于2016年10月在中国农业大学小麦遗传育种温室种植, 植株长至二叶一心时, 取小麦叶片, 按Saghai-Maroof等[11]的CTAB法提取基因组DNA。1.2 基因型分析
利用中国农业科学院作物科学研究所小麦基因组与基因资源研究团队开发的小麦55K SNP芯片对188个品种进行全基因组扫描。小麦55K芯片包含55,000个左右SNP标记, 均匀分布在小麦21条染色体上, 每条染色体平均有2600个左右SNP, 标记间的平均遗传距离约为0.1 cM, 平均物理距离小于300 kb, 并具有完整而详细的基因组信息。1.3 数据分析
针对SNP芯片的原始扫描结果, 利用Affymetrix的Axiom Analysis Suite软件分型。选择分型成功率(Call rate)>0.8、缺失率<10%、最小等位基因频率(MAF)>0.05过滤的50,743个SNP位点, 利用PowerMarker V3.25计算多态信息含量(PIC)和品种之间的遗传距离Nei[12]。根据Nei的遗传距离, 通过MEGA6软件(http://www.megasoftware.net/)构建遗传聚类树状图。2 结果与分析
2.1 多态性SNP位点的分布
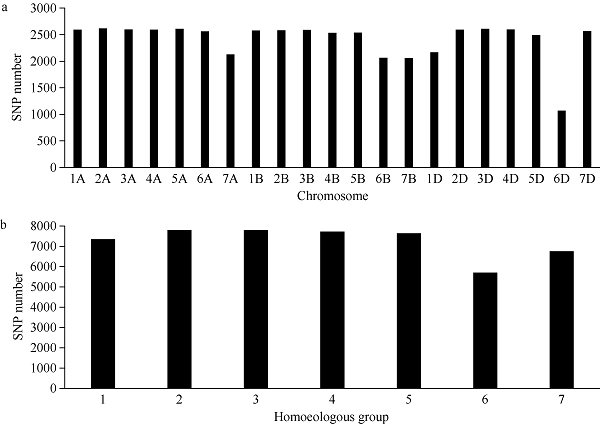
在分布于21条染色体的53,063个SNP位点中, 共计有50,743个SNP位点检测到多态性, 多态性比率为95.62% (50,743/53,063)。每条染色体分布1068~2616个多态性位点, 6D染色体上最少, 2A染色体上最多, 平均每条染色体2416个(图1-a)。除6D外, 染色体间标记数量差异不大(图1-a)。SNP标记在A、B和D基因组间分布比较均匀, A基因组的标记数最多, 有17,702个, 占34.88%; B基因组居中, 有16,943个, 占33.39%; D基因组的标记数最少, 有16,098个, 占31.73%。多态性SNP位点在7个部分同源群中的分布数量在5696~7797之间, 第 3部分同源群最多, 第6部分同源群最少, 分布从多到少依次为3>2>4>5>1>7>6 (图1-b)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1多态性位点在染色体(a)和部分同源群(b)上的分布
Fig. 1Distributions of polymorphic SNPs on individual chromosome (a) and each homoeologous group (b)
2.2 SNP标记多态性信息含量及品种间的遗传距离
基于SNP标记数据计算品种的多态性信息含量(polymorphic information content, PIC)值, 变异范围在0.090~0.375之间, 平均为0.265。134个地方品种的PIC值平均为0.229, 最高0.374, 最低0.090; 54个育成品种的PIC值平均为0.299, 最高0.375, 最低0.099 (表1)。Table 1
表1
表1地方品种和育成品种遗传差异比较
Table 1
| 类型 Type | 多态性信息含量 Polymorphism information content | 遗传距离 Genetic distance | |||||
|---|---|---|---|---|---|---|---|
| 平均值 Mean | 最大值 Max. value | 最小值 Min. value | 平均值 Mean | 最大值 Max. value | 最小值 Min. value | ||
| 地方品种 Landraces | 0.229 | 0.374 | 0.090 | 0.070 | 0.400 | 0.002 | |
| 育成品种 Cultivars | 0.299 | 0.375 | 0.099 | 0.114 | 0.337 | 0.004 | |
| 地方品种与育成品种之间 Between landraces and cultivars | 0.265 | 0.375 | 0.090 | 0.699 | 0.723 | 0.605 | |
新窗口打开|下载CSV
利用SNP标记数据计算188个品种两两之间的遗传距离, 共获得17,580个遗传距离值, 范围在0.002~0.723之间, 平均为0.378 (表1)。巴州库尔勒市的名为阿克库斯格的地方品种与巴州若羌县的名为冬麦的地方品种之间的遗传距离最小(0.002), 喀什地区莎车县的地方品种白芒白冬麦与育成品种新冬7号之间的遗传距离最大(0.723)。
按地方品种、育成品种、地方品种与育成品种之间3组比较遗传距离(表1), 134个地方品种两两之间的遗传距离值在0.002~0.400之间, 平均为0.070, 巴州库尔勒市的名为阿克库斯格的地方品种与巴州若羌县的名为冬麦的地方品种之间的遗传距离最小(0.002), 博乐市的地方品种小红芒与和田市的地方品种卡衣木江之间的遗传距离最大(0.400)。134个地方品种中, 有26个来源地不同、品种名皆为“白冬麦”的地方品种, 其中7个来自北疆各地区, 2个来自东疆的哈密地区, 17个来自南疆各地区。26个“白冬麦”之间的遗传距离在0.003~0.263之间, 2个分别来自乌鲁木齐市与昌吉州呼图壁县的白冬麦之间的遗传距离最小, 实际这2个品种的来源地之间的地理距离也较小; 2个分别来自北疆昌吉州阜康县与南疆喀什地区叶城县的白冬麦之间的遗传距离最大, 实际这2个品种来源地之间的地理距离也较大。
54个育成品种两两之间的遗传距离值在0.004~ 0.337之间, 平均为0.114, 新冬11号与新冬12号之间的遗传距离最小, 巴冬1号与新冬20号之间的遗传距离最大。由系谱可知, 新冬11号与新冬12号的母本均为杂交组合热衣木夏/亥恩·亥德; 巴冬1号母本是新疆的冬小麦地方品种阿克库斯格, 父本是意大利引进冬小麦品种奥维斯特, 而新冬20是由河北省农林科学院育成的冀87-5018系选而来。
134个地方品种与54个育成品种之间的遗传距离在0.605~0.723之间, 平均为0.699, 说明新疆冬小麦地方品种与育成品种之间的遗传差异大于地方品种之间的遗传差异, 也大于育成品种之间的遗传差异。伊宁市的地方品种无芒麦与育成品种奎冬5号之间的遗传距离最小(0.605), 喀什地区莎车县的地方品种白芒白冬麦与育成品种新冬7号之间的遗传距离最大(0.723)。
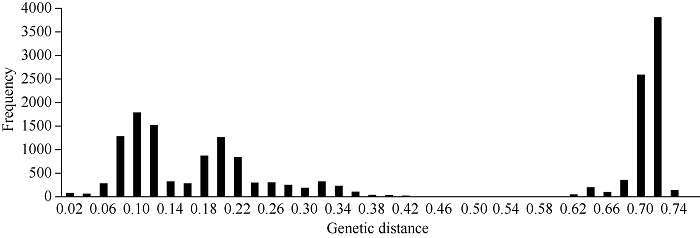
以0.02为组距进行次数分布频数分析(图2), 188个品种间的遗传距离分布频数呈现2个波峰曲线, 一个是地方小麦品种之间以及育成品种之间的遗传距离分布频数曲线, 在0.1处密度最大, 主要分布在0.06~0.12之间, 占26.10% (4588/17,580); 另一个是地方品种与育成品种之间的遗传距离分布频数曲线, 在0.72处分布密度最大, 主要分布在0.68~ 0.72之间, 占36.39% (6397/17,580)。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2品种间遗传距离的次数分布
Fig.2Distribution of genetic distances among wheat landraces and cultivars
2.4 聚类分析
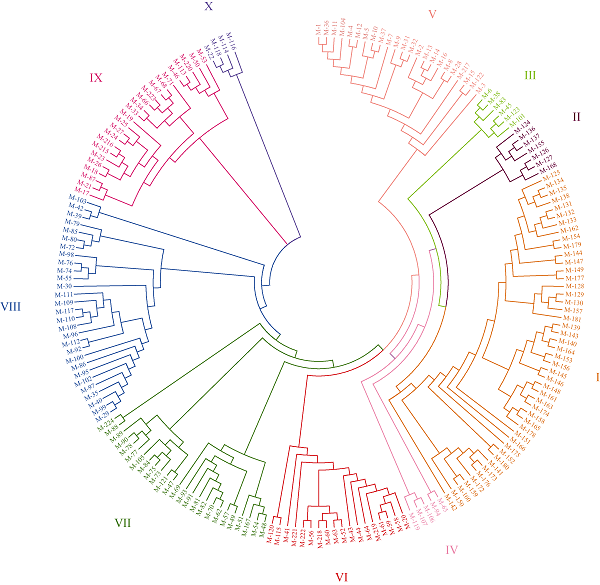
根据所有小麦品种的SNP, 利用PowerMarker V3.25 得到188个小麦材料两两之间的遗传距离矩阵, 并构建了NJ聚类图(图3)。通过聚类, 我们将所有小麦材料分为10个不同类群, 其中第I类(47个品种)和第II类(7个品种)均为育成品种, 其余类群均为地方品种, 其中第III类有6个品种, 第IV类有5个品种, 第V类23个品种, 第VI类18个品种, 第VII类25个品种, 第VIII类29个品种, 第IX类24个品种, 第X类最少, 仅有4个品种。各类群及包含的小麦材料详见表3及表4。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3188个新疆小麦地方品种和育成品种的SNP数据聚类图
代码的含义同
Fig. 3Phylogenetic tree of 188 wheat varieties in Xinjiang based on SNP
Codes correspond with those given in
Table 3
表3
表3各类群小麦品种列表
Table 3
| 类群 Group | 品种个数 Number of variety | 亚类 Sub-group | 品种名称 Variety name |
|---|---|---|---|
| I | 47 | 1 | 新冬11号, 新冬12号, 巴冬3号, 新冬1号, 新冬7号, 新冬9号, 新冬10号, 昌冬5号, 石冬5号, 新冬26号, 新冬14号, 新冬17号, 新冬19号, 新冬38号, 新冬5号, 新冬6号, 奎冬4号, 新冬4号, 新冬32号 Xindong 11, Xindong 12, Badong 3, Xindong 1, Xindong 7, Xindong 9, Xindong 10, Changdong 5, Shidong 5, Xindong 26, Xindong 14, Xindong 17, Xindong 19, Xindong 38, Xindong 5, Xindong 6, Kuidong 4, Xindong 4, Xindong 32 |
| 2 | 喀冬1号, 伊农3号, 喀冬3号, 喀冬5号, 石冬4号, 奎花1号, 新冬15号, 新冬16号, 新冬18号, 原冬3号, 塔冬5号, 伊农20号, 奎冬5号, 新冬22号, 伊农61号 Kadong 1, Yinong 3, Kadong 3, Kadong 5, Shidong 4, Kuihua 1, Xindong 15, Xindong 16, Xindong 18, Yuandong 3, Tadong 5, Yinong 20, Kuidong 5, Xindong 22, Yinong 61 | ||
| 3 | 新冬23号, 新冬24号, 伊农21号, 新冬25号, 新冬46号, 伊农18号, 新冬29号, 喀冬4号, 伊农16号, 黄河1号, 黄河3号, 新冬20号, 伊农1号 Xindong 23, Xindong 24, Yinong 21, Xindong 25, Xindong 46, Yinong 18, Xindong 29, Kadong 4, Yinong 16, Huanghe 1, Huanghe 3, Xindong 20, Yinong 1 | ||
| II | 7 | 新冬2号, 新冬3号, 新冬13号, 巴冬1号, 巴冬2号, 喀冬2号, 石冬6号 Xindong 2, Xindong 3, Xindong 13, Badong 1, Badong 2, Kadong 2, Shidong 6 | |
| III | 6 | 无芒麦(M-6), 无芒麦(M-38), 红冬麦(M-83), 南疆冬麦, 卡衣木江, 白晚冬麦(M-101) Wumangmai (M-6), Wumangmai (M-38), Hongdongmai (M-83), Nanjiangdongmai, Kayimujiang, Baiwandongmai (M-101) | |
| IV | 5 | 红冬麦(M-65), 安迪然冬麦, 白冬麦(M-106), 白冬麦(M-107), 黑冬麦(M-119) Hongdongmai (M-65), Andirandongmai, Baidongmai (M-106), Baidongmai (M-107), Heidongmai (M-119) | |
| V | 23 | 白冬麦(M-1), 晚熟冬麦, 白冬麦(M-11), 白冬麦(M-104), 白冬麦(M-4), 小白冬麦(M-12), 红壳白粒红冬麦, 红冬麦(M-10), 阿尔泰冬麦, 红壳冬麦(M-7), 白冬麦(M-31), 小冬麦(M-32), 白冬麦(M-9), 红壳小白冬麦, 青心兰麦, 红冬麦(M-16), 红冬麦(M-2), 红冬麦(M-8), 小红芒, 鄯善红冬麦, 红冬麦(M-15), 白冬麦(M-3), 白冬麦(M-122) Baidongmai (M-1), Wanshudongmai, Baidongmai (M-11), Baidongmai (M-104), Baidongmai (M-4), Xiaobaidongmai (M-12), Hongkebailihongdongmai, Hongdongmai (M-10), Aertaidongmai, Hongkedongmai (M-7), Baidongmai (M-31), Xiaodongmai (M-32), Baidongmai (M-9), Hongkexiaobaidongmai, Qingxinlanmai, Hongdongmai (M-16), Hongdongmai (M-2), Hongdongmai (M-8), Xiaohongmang, Shanshanhongdongmai, Hongdongmai (M-15), Baidongmai (M-3), Baidongmai (M-122) | |
| VI | 18 | 1 | 红穗白粒(M-20), 白冬麦(M-58), 白冬麦(M-59), 拉瓦得朗, 焉耆白冬麦, 尉力小麦, 南疆白冬麦, 红冬麦(M-44) Hongsuibaili (M-20), Baidongmai (M-58), Baidongmai (M-59), Lawadelang, Yanqibaidongmai, Yulixiaomai, Nanjiangbaidongmai, Hongdongmai (M-44) |
| 2 | 红冬麦(M-52), 沙雅冬麦(M-63), 热衣木夏, 和静红冬麦, 买洛瓦西(M-56), 库车买洛瓦西 Hongdongmai (M-52), Shayadongmai (M-63), Reyimuxia, Hejinghongdongmai, Mailuowaxi (M-56), Kuchemailuowaxi | ||
| 3 | 库尔勒白冬麦, 白冬麦(M-41), 白冬麦(M-115), 黑冬麦(M-120) Korlabaidongmai, Baidongmai (M-41), Baidongmai (M-115), Heidongmai (M-120) | ||
| VII | 25 | 1 | 阿克古斯格, 冬麦(M-54), 河西巴克红麦, 阿克库孜盖 Akegusige, Dongmai (M-54), Hexibakehongmai, Akekuzigai |
| 类群 Group | 品种个数 Number of variety | 亚类 Sub-group | 品种名称 Variety name |
| VII | 2 | 阳霞红冬麦, 纳瓦提然, 梁山冬麦, 沙雅冬麦, 阿克不大衣, 可孜不大衣, 白冬麦(M-91), 哈热克巴斯曼, 黑冬麦(M-69) Yangxiahongdongmai, Nawatiran, Liangshandongmai, Shayadongmai, Akebudayi, Kezibudayi, Baidongmai (M-91), Harekebasman, Heidongmai (M-69) | |
| 3 | 买洛瓦西(M-47), 早熟冬麦, 白冬麦(M-73), 白穗子, 土种麦, 黑穗麦, 白冬麦(M-78), 白冬麦(M-90), 白冬麦(M-77), 红皮红冬麦(M-89), 黑冬麦(M-88), 巴楚切利麦 Mailuowaxi (M-47), Zaoshudongmai, Baidongmai (M-73), Baisuizi, Tuzhongmai, Heisuimai, Baidongmai (M-78), Baidongmai (M-90), Baidongmai (M-77), Hongpihongdongmai (M-89), Heidongmai (M-88), Bachuqielimai | ||
| VIII | 28 | 1 | 白壳白粒红心冬麦, 长巴什曼, 红冬麦(M-110), 卡尔库孜干, 白冬麦(M-108), 黑冬麦(M-109), 哈阿热托司曼, 麦盖提白冬麦, 红冬麦(M-30) Baikebailihongxindongmai, Changbashenman, Hongdongmai (M-110), Kaerkuzigan, Baidongmai (M-108), Heidongmai (M-109), Haaretuosiman, Maigaitibaidongmai, Hongdongmai (M-30) |
| 2 | 白冬麦(M-29), 红芒安江, 红冬麦(M-40), 早熟冬麦(M-35), 白粒早熟, 红芒早, 乌恰白麦, 阿伯格其力格, 白芒白冬麦 Baidongmai (M-29), Hongmang’anjiang, Hongdongmai (M-40), Zaoshudongmai (M-35), Bailizaoshu, Hongmangzao, Wuqiabaimai, Abogeqilige, Baimangbaidongmai | ||
| 3 | 哈热克, 吧子条播, 冬麦(M-55), 白晚冬麦(M-98), 沙雅冬麦(M-70), 可孜不大衣, 白冬麦(M-85), 黑冬麦(M-79), 红冬麦(M-39), 焉耆冬麦, 白穗早熟冬麦 Hareke, Bazitiaobo, Dongmai (M-55), Baiwandongmai (M-98), Shayadongmai (M-70), Kezibudayi, Baidongmai (M-85), Heidongmai (M-79), Hongdongmai (M-39), Yanqidongmai, Baisuizaoshudongmai | ||
| IX | 24 | 1 | 昌吉小红冬麦, 玛纳斯红冬麦, 白芒冬麦, 红冬麦(M-18), 红冬麦(M-26), 白穗白粒, 白冬麦(M-87), 红穗粒(M-17), 金包银(M-24), 白冬麦(M-27), 红壳冬麦(M-25), 红穗白粒(M-19) Changjixiaohongdongmai, Manasihongdongmai, Baimangdongmai, Hongdongmai (M-18), Hongdongmai (M-26), Baisuibaili, Baidongmai (M-87), Hongsuili (M-17), Jinbaoyin (M-24), Baidongmai (M-27), Hongkedongmai (M-25), Hongsuibaili (M-19) |
| 2 | 红冬麦(M-66), 沙雅冬麦(M-223), 红冬麦(M-67), 黑冬麦(M-68), 冬麦(M-33), 本地冬麦, 白冬麦(M-71) Hongdongmai (M-66), Shayadongmai (M-223), Hongdongmai (M-67), Heidongmai (M-68), Dongmai (M-33), Bendidongmai, Baidongmai (M-71) | ||
| 3 | 买洛瓦西(M-46), 黑冬麦(M-113), 尉力小麦, 巴利巴式, 白冬麦(M-53) Mailuowaxi (M-46), Heidongmai (M-113), Yulixiaomai, Balibashi, Baidongmai (M-53) | ||
| X | 4 | 白冬麦(M-22), 克孜尔库孜干, 黑穗冬麦, 黑冬麦(M-116) Baidongmai (M-22), Kezierkuzigan, Heisuidongmai, Heidongmai (M-116) |
新窗口打开|下载CSV
Table 4
表4
表4SNP分析的新疆冬小麦品种名称及来源
Table 4
| 序号 No. | 品种编号 Variety code | 品种名称 Variety name | 品种类型 Variety type | 来源地/系谱 Origin/pedigree |
|---|---|---|---|---|
| 1 | M-1 | 白冬麦 Baidongmai | 地方品种 Landrace | 乌鲁木齐市Urumqi |
| 2 | M-2 | 红冬麦 Hongdongmai | 地方品种 Landrace | 昌吉市Changji |
| 3 | M-3 | 白冬麦 Baidongmai | 地方品种 Landrace | 昌吉市Changji |
| 4 | M-4 | 白冬麦 Baidongmai | 地方品种 Landrace | 昌吉市Changji |
| 序号 No. | 品种编号 Variety code | 品种名称 Variety name | 品种类型 Variety type | 来源地/系谱 Origin/pedigree |
| 5 | M-5 | 红壳白粒红冬麦 Hongkebailihongdongmai | 地方品种 Landrace | 昌吉州米泉县Miquan, Changji |
| 6 | M-6 | 无芒麦 Wumangmai | 地方品种 Landrace | 昌吉州米泉县Miquan, Changji |
| 7 | M-7 | 红壳冬麦 Hongkedongmai | 地方品种 Landrace | 昌吉州米泉县Miquan, Changji |
| 8 | M-8 | 红冬麦 Hongdongmai | 地方品种 Landrace | 昌吉州奇台县Qitai, Changji |
| 9 | M-9 | 白冬麦 Baidongmai | 地方品种 Landrace | 昌吉州阜康县Fukang, Changji |
| 10 | M-10 | 红冬麦 Hongdongmai | 地方品种 Landrace | 昌吉州阜康县Fukang, Changji |
| 11 | M-11 | 白冬麦 Baidongmai | 地方品种 Landrace | 昌吉州呼图壁县Hutubi, Changji |
| 12 | M-12 | 小白冬麦 Xiaobaidongmai | 地方品种 Landrace | 昌吉州呼图壁县Hutubi, Changji |
| 13 | M-13 | 红壳小白冬麦 Hongkexiaobaidongmai | 地方品种 Landrace | 昌吉州玛纳斯县Manasi, Changji |
| 14 | M-14 | 青心兰麦 Qingxinlanmai | 地方品种 Landrace | 昌吉州玛纳斯县Manasi, Changji |
| 15 | M-15 | 红冬麦 Hongdongmai | 地方品种 Landrace | 昌吉州玛纳斯县Manasi, Changji |
| 16 | M-16 | 红冬麦 Hongdongmai | 地方品种 Landrace | 昌吉州玛纳斯县Manasi, Changji |
| 17 | M-17 | 红穗粒 Hongsuili | 地方品种 Landrace | 吐鲁番市Turpan |
| 18 | M-18 | 红冬麦 Hongdongmai | 地方品种 Landrace | 吐鲁番市Turpan |
| 19 | M-19 | 红穗白粒 Hongsuibaili | 地方品种 Landrace | 吐鲁番市Turpan |
| 20 | M-20 | 红穗白粒 Hongsuibaili | 地方品种 Landrace | 吐鲁番市Turpan |
| 21 | M-21 | 白穗白粒 Baisuibaili | 地方品种 Landrace | 吐鲁番市Turpan |
| 22 | M-22 | 白冬麦 Baidongmai | 地方品种 Landrace | 哈密市Hami |
| 23 | M-23 | 白芒冬麦 Baimangdongmai | 地方品种 Landrace | 哈密市Hami |
| 24 | M-24 | 金包银 Jinbaoyin | 地方品种 Landrace | 哈密市Hami |
| 25 | M-25 | 红壳冬麦 Hongkedongmai | 地方品种 Landrace | 哈密市Hami |
| 26 | M-26 | 红冬麦 Hongdongmai | 地方品种 Landrace | 哈密市Hami |
| 27 | M-27 | 白冬麦 Baidongmai | 地方品种 Landrace | 哈密市Hami |
| 28 | M-28 | 小红芒 Xiaohongmang | 地方品种 Landrace | 博乐市Bole |
| 29 | M-29 | 白冬麦 Baidongmai | 地方品种 Landrace | 塔城市Tacheng |
| 30 | M-30 | 红冬麦 Hongdongmai | 地方品种 Landrace | 塔城市Tacheng |
| 31 | M-31 | 白冬麦 Baidongmai | 地方品种 Landrace | 塔城地区沙湾县Shawan, Tacheng |
| 32 | M-32 | 小冬麦 Xiaodongmai | 地方品种 Landrace | 塔城地区沙湾县Shawan, Tacheng |
| 33 | M-33 | 冬麦 Dongmai | 地方品种 Landrace | 塔城地区乌苏县Wusu, Tacheng |
| 34 | M-34 | 本地冬麦 Bendidongmai | 地方品种 Landrace | 塔城地区乌苏县Wusu, Tacheng |
| 35 | M-35 | 早熟冬麦 Zaoshudongmai | 地方品种 Landrace | 阿勒泰地区布尔津县Buerjing, Aletai |
| 36 | M-36 | 晚熟冬麦 Wanshudongmai | 地方品种 Landrace | 阿勒泰地区布尔津县Buerjing, Aletai |
| 37 | M-37 | 阿尔泰冬麦 Aertaidongmai | 地方品种 Landrace | 阿勒泰地区布尔津县Buerjing, Aletai |
| 38 | M-38 | 无芒麦 Wumangmai | 地方品种 Landrace | 伊宁市Yining |
| 39 | M-39 | 红冬麦 Hongdongmai | 地方品种 Landrace | 巴州和静县Hejing, Bazhou |
| 40 | M-40 | 红冬麦 Hongdongmai | 地方品种 Landrace | 巴州和静县Hejing, Bazhou |
| 41 | M-41 | 白冬麦 Baidongmai | 地方品种 Landrace | 巴州和静县Hejing, Bazhou |
| 42 | M-42 | 焉耆冬麦 Yanqidongmai | 地方品种 Landrace | 巴州焉耆县Yanqi, Bazhou |
| 43 | M-43 | 南疆白冬麦Nanjiangbaidongmai | 地方品种 Landrace | 巴州焉耆县Yanqi, Bazhou |
| 44 | M-44 | 红冬麦 Hongdongmai | 地方品种 Landrace | 巴州焉耆县 Yanqi, Bazhou |
| 45 | M-45 | 南疆冬麦 Nanjiangdongmai | 地方品种 Landrace | 巴州焉耆县Yanqi, Bazhou |
| 46 | M-46 | 麦洛瓦西 Mailuowaxi | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 47 | M-47 | 麦洛瓦西 Mailuowaxi | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 48 | M-48 | 阿克库斯格 Akegusige | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 49 | M-49 | 阳霞红冬麦Yangxiahongdongmai | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 50 | M-50 | 巴利巴式 Balibashi | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 51 | M-51 | 阿克库孜盖 Akekuzigai | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 52 | M-52 | 红冬麦 Hongdongmai | 地方品种 Landrace | 库尔勒市Korla, Bazhou |
| 53 | M-53 | 白冬麦 Baidongmai | 地方品种 Landrace | 巴州若羌县Ruoqiang, Bazhou |
| 54 | M-54 | 冬麦 Dongmai | 地方品种 Landrace | 巴州若羌县Ruoqiang, Bazhou |
| 55 | M-55 | 冬麦 Dongmai | 地方品种 Landrace | 巴州轮台县Luntai, Bazhou |
| 56 | M-56 | 买洛瓦西 Mailuowaxi | 地方品种 Landrace | 阿克苏地区库车县Kuche, Aksu |
| 57 | M-57 | 纳瓦提然 Nawatiran | 地方品种 Landrace | 阿克苏地区新和县Xinhe, Aksu |
| 58 | M-58 | 白冬麦 Baidongmai | 地方品种 Landrace | 阿克苏地区新和县Xinhe Aksu |
| 59 | M-59 | 白冬麦 Baidongmai | 地方品种 Landrace | 阿克苏地区新和县Xinhe, Aksu |
| 60 | M-60 | 热衣木夏 Reyimuxia | 地方品种 Landrace | 阿克苏地区新和县Xinhe, Aksu |
| 61 | M-61 | 拉瓦得朗 Lawadelang | 地方品种 Landrace | 阿克苏地区沙雅县Shaya, Aksu |
| 62 | M-62 | 梁山冬麦 Liangshandongmai | 地方品种 Landrace | 阿克苏地区新和县Kuche, Aksu |
| 63 | M-63 | 沙雅冬麦 Shayadongmai | 地方品种 Landrace | 阿克苏地区沙雅县Shaya, Aksu |
| 64 | M-64 | 尉力小麦 Weilixiaomai | 地方品种 Landrace | 库尔勒市Korla |
| 65 | M-65 | 红冬麦 Hongdongmai | 地方品种 Landrace | 阿克苏地区温宿县Wensu, Aksu |
| 66 | M-66 | 红冬麦 Hongdongmai | 地方品种 Landrace | 阿克苏市Akesu |
| 67 | M-67 | 红冬麦 Hongdongmai | 地方品种 Landrace | 阿克苏市Akesu |
| 68 | M-68 | 黑冬麦 Heidongmai | 地方品种 Landrace | 阿克苏地区阿瓦提县Awati, Aksu |
| 69 | M-69 | 黑冬麦 Heidongmai | 地方品种 Landrace | 阿克苏地区阿瓦提县Awati, Aksu |
| 70 | M-70 | 沙雅冬麦 Shayadongmai | 地方品种 Landrace | 阿克苏地区阿瓦提县Awati, Aksu |
| 71 | M-71 | 白冬麦 Baidongmai | 地方品种 Landrace | 阿克苏地区阿瓦提县Awati, Aksu |
| 72 | M-72 | 红皮冬麦 Hongpidongmai | 地方品种 Landrace | 阿克苏地区乌什县Wushi, Aksu |
| 73 | M-73 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 74 | M-74 | 哈热克 Hareke | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 75 | M-75 | 白穗子 Baisuizi | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 76 | M-76 | 吧子条播 Bazitiaobo | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 77 | M-77 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 78 | M-78 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区伽师县Jiashi, Kashi |
| 79 | M-79 | 黑冬麦 Heidongmai | 地方品种 Landrace | 阿克苏地区库车县Kuche, Aksu |
| 80 | M-80 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区疏附县Shufu, Kashi |
| 81 | M-81 | 阿克不大衣 Akebudayi | 地方品种 Landrace | 喀什地区疏附县Shufu, Kashi |
| 82 | M-82 | 可孜不大衣 Kezibudayi | 地方品种 Landrace | 喀什地区疏附县Shufu, Kashi |
| 83 | M-83 | 红冬麦 Hongdongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 84 | M-84 | 土种麦 Tuzhongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 85 | M-85 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 86 | M-86 | 阿伯格其力格 Abogqilige | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 87 | M-87 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 88 | M-88 | 黑冬麦 Heidongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 89 | M-89 | 红皮红冬麦 Hongpihongdongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 90 | M-90 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区疏勒县Shule, Kashi |
| 91 | M-91 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什市Kashi |
| 92 | M-92 | 白壳白粒红心冬麦 Baikebailihongxindongmai | 地方品种 Landrace | 喀什市Kashi |
| 93 | M-93 | 哈热克巴斯曼Harekebasman | 地方品种 Landrace | 喀什市Kashi |
| 94 | M-94 | 安迪然冬麦 Andirandongmai | 地方品种 Landrace | 喀什市Kashi |
| 95 | M-95 | 乌恰白麦 Wuqiabaimai | 地方品种 Landrace | 阿克苏地区乌恰县Wuqia, Aksu |
| 96 | M-96 | 麦盖提白冬麦Maigaitibaidongmai | 地方品种 Landrace | 喀什地区麦盖提县Maigaiti, Kashi |
| 97 | M-97 | 白粒早熟 Bailizaoshu | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 98 | M-98 | 白晚冬麦 Baiwandongmai | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 99 | M-99 | 红芒安江 Hongmang’anjiang | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 100 | M-100 | 白芒白冬麦Baimangbaidongmai | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 101 | M-101 | 白晚冬麦 Baiwandongmai | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 102 | M-102 | 红芒早 Hongmangzao | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 103 | M-103 | 白穗早熟冬麦Baisuizaoshudongmai | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 104 | M-104 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区莎车县Shache, Kashi |
| 105 | M-105 | 黑穗麦 Heisuimai | 地方品种 Landrace | 喀什地区泽普县Zepu, Kashi |
| 106 | M-106 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 107 | M-107 | 白冬麦 Baidongmai | 地方品种 Landrace | 和田地区洛浦县Luopu, Hetian |
| 108 | M-108 | 白冬麦 Baidongmai | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 109 | M-109 | 黑冬麦 Heidongmai | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 110 | M-110 | 红冬麦 Hongdongmai | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 111 | M-111 | 哈阿热托司曼Haaretuosiman | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 112 | M-112 | 长巴什曼 Changbashenman | 地方品种 Landrace | 喀什地区叶城县Yecheng, Kashi |
| 113 | M-113 | 黑冬麦 Heidongmai | 地方品种 Landrace | 和田地区墨玉县Moyu, Hetian |
| 114 | M-114 | 黑穗冬麦 Heisuidongmai | 地方品种 Landrace | 和田地区洛浦县Luopu, Hetian |
| 115 | M-115 | 白冬麦 Baidongmai | 地方品种 Landrace | 和田地区洛浦县Luopu, Hetian |
| 116 | M-116 | 黑冬麦 Heidongmai | 地方品种 Landrace | 和田地区洛浦县Luopu, Hetian |
| 117 | M-117 | 卡尔库孜干 Kaerkuzigan | 地方品种 Landrace | 和田地区于田县Yutian, Hetian |
| 118 | M-118 | 克孜尔库孜干 Kezierkuzigan | 地方品种 Landrace | 和田地区民丰县Minfeng, Hetian |
| 119 | M-119 | 黑冬麦 Heidongmai | 地方品种 Landrace | 和田市Hetian |
| 120 | M-120 | 黑冬麦 Heidongmai | 地方品种 Landrace | 和田市Hetian |
| 121 | M-121 | 早熟冬麦 Zaoshudongmai | 地方品种 Landrace | 和田市Hetian |
| 122 | M-122 | 白冬麦 Baidongmai | 地方品种 Landrace | 和田市Hetian |
| 123 | M-123 | 卡衣木江 Kayimujiang | 地方品种 Landrace | 和田市Hetian |
| 124 | M-167 | 河西巴克红麦Hexibakehongmai | 地方品种 Landrace | 库尔勒市Korla |
| 125 | M-215 | 昌吉小红冬麦Changjixiaohongdongmai | 地方品种 Landrace | 昌吉市Changji |
| 126 | M-216 | 玛纳斯红冬麦Manasihongdongmai | 地方品种 Landrace | 昌吉州玛纳斯县Manasi, Changji |
| 127 | M-217 | 鄯善红冬麦Shanshanhongdongmai | 地方品种 Landrace | 吐鲁番地区鄯善县Shanshan, Turpan |
| 128 | M-218 | 和静红冬麦Hejing hong dong mai | 地方品种 Landrace | 巴州和静县Hejing, Bazhou |
| 129 | M-219 | 焉耆白冬麦Yanqibaidongmai | 地方品种 Landrace | 阿克苏地区焉耆县Yanqi, Aksu |
| 130 | M-220 | 尉力小麦 Yulixiaomai | 地方品种 Landrace | 巴州尉犁县Yuli, Bazhou |
| 131 | M-221 | 库尔勒白冬麦Korlabaidongmai | 地方品种 Landrace | 库尔勒市Korla |
| 132 | M-222 | 库车麦洛瓦西Kuchemailuowaxi | 地方品种 Landrace | 阿克苏地区库车县Kuche, Aksu |
| 133 | M-223 | 沙雅冬麦 Shayadongmai | 地方品种 Landrace | 和田地区沙雅县Shaya, Hetian |
| 134 | M-224 | 巴楚切利麦 Bachuqielimai | 地方品种 Landrace | 喀什地区巴楚县Bachu, Kasha |
| 135 | M-124 | 喀冬2号 Kadong 2 | 育成品种 Cultivar | 乌克兰0246/叶城春麦 Ukraine 0246/Yechengchunmai |
| 136 | M-125 | 新冬1号 Xindong 1 | 育成品种 Cultivar | 乌克兰0246/阿克苏红冬麦//敖德萨3号 Ukraine 0246/Aksuhongdongmai// Aodesa 3 |
| 137 | M-126 | 新冬2号 Xindong 2 | 育成品种 Cultivar | 热衣木夏/亥恩·亥德3个F5选系混合 Three F5 selection combinations from Reyimuxia/Heine Hvede |
| 138 | M-127 | 新冬3号 Xindong 3 | 育成品种 Cultivar | 阿克苏红冬麦/亥恩·亥德 Aksuhongdongmai/Heine Hvede |
| 139 | M-128 | 新冬4号 Xindong 4 | 育成品种 Cultivar | 巴克甫克/北京7号 Bakefuke/Beijing 7 |
| 140 | M-129 | 新冬5号Xindong 5 | 育成品种 Cultivar | 巴克甫克/北京7号 Bakefuke/Beijing 7 |
| 141 | M-130 | 新冬6号 Xindong 6 | 育成品种 Cultivar | 新冬2号姊妹系/奥德萨3号 Xindong 2 sib line/Aodesa 3 |
| 142 | M-131 | 新冬7号 Xindong 7 | 育成品种 Cultivar | 新冬2号姊妹系/奥德萨3号 Xindong 2 sib line/Odessa 3 |
| 143 | M-132 | 新冬9号 Xindong 9 | 育成品种 Cultivar | 热衣木夏/亥恩·亥德 Reyimuxia/Heine Hvede |
| 144 | M-133 | 新冬10号 Xindong 10 | 育成品种 Cultivar | 新冬2号/奥德萨3号 Xindong 2/Aodesa 3 |
| 145 | M-134 | 新冬11号 Xindong 11 | 育成品种 Cultivar | 白沟头(热衣木夏/亥恩·亥德)//2407 (巴克甫克/北京7号) Baigoutou (Reyimuxia/Heine Hvede)//2407 (Bakefuke/Beijing 7) |
| 146 | M-135 | 新冬12号 Xindong 12 | 育成品种 Cultivar | 新冬2号//2407(新冬2号/奥德萨3号) Xindong 2//2407 (Xindong 2/Aodesa 3) |
| 147 | M-136 | 巴冬1号 Badong 1 | 育成品种 Cultivar | 阿克库斯格/奥维斯特(意大利) Akekusige/Ovist (Italy) |
| 148 | M-137 | 巴冬2号 Badong 2 | 育成品种 Cultivar | 巴克甫克/奥维斯 Bakefuke/Ovist |
| 149 | M-138 | 巴冬3号 Badong 3 | 育成品种 Cultivar | 从华北497中选单株育成 Individual plant selection from Huabei 497 |
| 150 | M-139 | 喀冬1号 Kadong 1 | 育成品种 Cultivar | 巴克甫克/亥恩·亥德 Bakefuke/Heine Hvede |
| 151 | M-140 | 喀冬3号 Kadong 3 | 育成品种 Cultivar | 自中农院3026 (早洋麦/鹅冠186)中选育成 Selected from CAAS 3026 (Zaoyang mai/Eguan 186) |
| 152 | M-141 | 喀冬4号 Kadong 4 | 育成品种 Cultivar | 从华北187中系统选育 Selected from Huabei 187 |
| 153 | M-142 | 伊农1号 Yinong 1 | 育成品种 Cultivar | 新乌克兰84/西北612 New Ukraine 84/Xibei 612 |
| 154 | M-143 | 伊农3号 Yinong 3 | 育成品种 Cultivar | 西北134/无芒4号 Xibei 134/Wumang 4 |
| 155 | M-144 | 新冬14号 Xindong 14 | 育成品种 Cultivar | 热衣木夏/新乌克兰83 Reyimuxia/New Ukraine 83 |
| 156 | M-145 | 新冬15号 Xindong 15 | 育成品种 Cultivar | 新冬2号/ 50周年(前苏联品种) Xindong 2/50 anniversary (Former Soviet Union) |
| 157 | M-146 | 新冬16号 Xindong 16 | 育成品种 Cultivar | (巴克甫克/亥恩·亥德)选系71-66/70-4 (新冬7号姊妹系) (Bakefuke/Heine Hvede) Selection 71-66/70-4 (Xindong 7 sib line) |
| 158 | M-147 | 新冬17号 Xindong 17 | 育成品种 Cultivar | 新冬14/河南安选5号 Xindong 14/Henan’anxuan 5 |
| 159 | M-148 | 新冬18号 Xindong 18 | 育成品种 Cultivar | NS11-33 (南斯拉夫品种)/新冬3号 NS11-33 (Yugoslavia variety)/Xindong 3 |
| 160 | M-149 | 新冬19号 Xindong 19 | 育成品种 Cultivar | 170/阿芙乐尔//H矮82-6 (组培品系)辐射选育 170/Aphrora//H short 82-6 (tissue culture line) Radiation selection |
| 161 | M-150 | 新冬20号 Xindong 20 | 育成品种 Cultivar | 原系号冀87-5018 (河北省农林科学院粮食作物研究所) Old system number Ji 87-5018 (Food Crop Research Institute, HAAS) |
| 162 | M-151 | 新冬23号 Xindong 23 | 育成品种 Cultivar | 美国引进的1个F2分离群体/自育花培品系88-136 An F2 segregation population from USA/88-136 |
| 163 | M-152 | 新冬25号 Xindong 25 | 育成品种 Cultivar | 冀885-443/冀88-5282 Ji 885-443/Ji 88-5282 |
| 164 | M-153 | 石冬4号 Shidong 4 | 育成品种 Cultivar | |
| 165 | M-154 | 石冬5号 Shidong 5 | 育成品种 Cultivar | 红选501/唐努尔(墨西哥春麦)//红选501/西埃代赛落斯(墨西哥春麦) Hongxuan 501/Tangnuer (Mexico)// Hongxuan 501/West edaselos (Mexico) |
| 166 | M-155 | 石冬6号 Shidong 6 | 育成品种 Cultivar | 春小麦京5029/冬小麦77-21 Spring wheat Jing 5029/Winter Wheat 77-21 |
| 167 | M-156 | 奎花1号 Kuihua 1 | 育成品种 Cultivar | 京花1号/奎冬3号 Jinghua 1/Kuidong 3 |
| 168 | M-157 | 奎冬4号 Kuihua 4 | 育成品种 Cultivar | (热衣木夏/亥恩·亥德) F3选系/奥德萨3号//7401-54/704/山前麦 (Reyimuxia/Heine Hvede) F3 selection/Aodesa 3//7401-54/704/Shanqianmai |
| 169 | M-158 | 奎冬5号 Kuihua 5 | 育成品种 Cultivar | 加拿大冬小麦品种Nostar/花春84-1//76-4/洛夫林13 Nostar (Canada winter wheat)/ Huachun 84-1//76-4/Lovrin 13 |
| 170 | M-159 | 黄河1号 Huanghe 1 | 育成品种 Cultivar | |
| 171 | M-160 | 黄河3号 Huanghe 3 | 育成品种 Cultivar | |
| 172 | M-161 | 原冬3号 Yuandong 3 | 育成品种 Cultivar | (冀麦2号/阿芙乐尔) γ射线处理 (Jimai 2/Aurora) γ-ray |
| 173 | M-162 | 昌冬5号 Changdong 5 | 育成品种 Cultivar | |
| 174 | M-163 | 塔冬5号 Tadong 5 | 育成品种 Cultivar | |
| 175 | M-164 | 喀冬5号 Kadong 5 | 育成品种 Cultivar | |
| 176 | M-165 | 新冬22号 Xindong 22 | 育成品种 Cultivar | 加拿大冬小麦品种Nostar/花春84-1// 76-4/洛夫林13 Canadian winter variety Nostar/ Huachun 84-1//76-4/Lovrin 13 |
| 177 | M-166 | 新冬24号 Xindong 24 | 育成品种 Cultivar | 9245/冀6159 9245/Ji 6159 |
| 178 | M-168 | 新冬13号 Xindong 13 | 育成品种 Cultivar | 新冬3号/乌克兰0246 Xindong 3/Ukraine 0246 |
| 179 | M-172 | 伊农16号 Yinong 16 | 育成品种 Cultivar | 白欧柔/72-629/K2-13//72-629/7141- 64/K2-13 White Orofen/72-629/K2-13//72-629/ 7141-64/K2-13 |
| 180 | M-173 | 伊农18号 Yinong 18 | 育成品种 Cultivar | 81283/81243 |
| 181 | M-174 | 伊农20号 Yinong 20 | 育成品种 Cultivar | 农大64015/8103-12-6-3 Nongda 64015/8103-12-6-3 |
| 182 | M-175 | 伊农21号 Yinong 21 | 育成品种 Cultivar | 西北612/乌克兰84/太谷核不育系/鲁麦13 Xibei 612/Ukraine 84/Taigu genic male-sterile line |
| 183 | M-176 | 新冬29号 Xindong 29 | 育成品种 Cultivar | PH82-2-2/鲁植79-1 PH82-2-2/Luzhi 79-1 |
| 184 | M-177 | 新冬38号 Xindong 29 | 育成品种 Cultivar | (矮秆916/9133) F1/伊农16 (Dwarf 916/9133) F1/Yinong 16 |
| 185 | M-178 | 伊农61号 Yinong 61 | 育成品种 Cultivar | 马苏托小麦/大头麦/Westmount//61-4 ///84114 Masutuo/ Datoumai/ Westmount//61-4///84114 |
| 186 | M-179 | 新冬26号 Xindong 26 | 育成品种 Cultivar | 红选501/唐努尔(墨西哥春麦)//红选501/西埃代赛洛斯(墨西哥春麦) Hongxuan 501/Tangnuer (Mexico)// Hongxuan 501/West edaselos (Mexico) |
| 187 | M-180 | 新冬46号 Xindong 46 | 育成品种 Cultivar | 农大3338/S180 Nongda 3338/S180 |
| 188 | M-181 | 新冬32号 Xindong 32 | 育成品种 Cultivar | 1998年从矮败轮回选择群体中选择 Selected from Aibai recurrent selection population in 1998 |
新窗口打开|下载CSV
第I类47个品种均为育成品种, 遗传距离在0.004~0.337之间, 分为3个亚类。第1亚类19个品种, 亲本多含有国外引进冬小麦品种, 如石冬6号与新冬26号, 均为中国科学院新疆生态与地理研究所选育的耐盐冬小麦品种, 且均以冬小麦品种红选501与墨西哥春麦品种唐努尔杂交的F1代为母本, 再与红选501和墨西哥春麦品种西埃代赛洛斯杂交的F1代为父本而育成的品种。最为突出的是新冬2号冬小麦品种, 是新疆农业科学院粮食作物研究所与新疆八一农学院(现新疆农业大学)以新疆冬小麦地方品种热衣木夏为母本、丹麦引进多花多实冬小麦品种亥恩·亥德为父本杂交选育而成, 于1966年审定命名[13]; 新疆农业科学院粮食作物研究所又以新冬2号姊妹系为母本、前苏联引进冬小麦品种奥德萨3号为父本, 进一步选育得到新冬6号、新冬7号、新冬10号3个品种。新冬号冬小麦品种曾在全疆推广种植, 对新疆小麦产量的提高起了很大作用, 其中新冬2号1980年种植23.3万公顷, 占全疆冬小麦面积的1/3。新疆农垦科学院以新疆冬小麦地方品种热衣木夏与前苏联引进品种新乌克兰83号杂交选育成新冬14号, 又利用新冬14号与河南安选5号杂交得到新冬17号小麦品种。第2亚类15个品种, 亲缘关系比较复杂, 其中奎冬5号与新冬22号均以加拿大引进小麦品种Nostar与花春84-1杂交F1为母本, 以高代品系76-4与引自罗马尼亚的洛夫林13杂交F1为父本选育而来; 喀冬1号、奎花1号、新冬15号、新冬16号、新冬18号均为利用我国新疆冬小麦地方品种(巴克甫克、热衣木夏、阿克苏红冬麦)与丹麦引进品种亥恩·亥德的杂交后代再与其他国内外冬小麦品种杂交选育而来, 如引自前苏联的无芒4号、小鹅186、阿芙乐尔、50周年等, 以及国内其他省份的西北134、华北187、京花1号、山东蚰包、冀麦2号等。第3亚类有13个品种, 均含有引自国内其他省份的冬小麦优良品种血缘, 如引自河北的冀6159、冀87-5018, 引自山东的PH82-2- 2、鲁植79-1, 引自陕西的西北612、山西的太谷核不育系以及庄巧生院士选育的优良品种华北187等。其中喀冬4号、新冬20号、新冬46号分别是华北187、冀87-5018、农大211引入新疆试种推广后审定的品种, 新冬25号的父母本均为河北的品种。
第II类7个品种均为育成品种, 遗传距离在0.006~0.269之间, 新冬2号与新冬3号之间的遗传距离最小, 喀冬2号与新冬13号之间的遗传距离最大。其中新冬2号与新冬3号的亲本之一都是上世纪50至60年代冬小麦育种多选的、引自丹麦的多粒源亲本亥恩·亥德, 新疆农业科学院粮食作物研究所又以品质表现较好的新冬3号为母本、较抗病抗寒的前苏联冬小麦品种乌克兰0246为父本进行杂交, 培育出了中产、品质好、容重高且抗病、抗寒性等都比新冬2号有所提高的新冬13号小麦品种。此亚类中巴州1号和巴州2号的共同亲本之一是引自意大利的冬小麦品种奥维斯特; 喀冬2号(乌克兰0246/叶城春麦)和石冬6号(春小麦京5029/辐射育种冬小麦品系77-21)的亲本之一都是春麦, 均具有早熟特性。
自第III类到第X类均为地方品种, 详述如下。
第III类有6个品种, 遗传距离在0.149~0.211之间。其中2份分别来自北疆昌吉州米泉县和伊犁州伊宁县的名称均无芒麦的品种聚在一起, 二者之间的遗传距离为0.166。其余4份材料均来自南疆地区, 2份来自喀什, 1份来自和田, 1份来自巴州, 地域相邻。
第IV类有5个品种, 遗传距离在0.093~0.274之间。2个分别来自喀什地区叶城县和和田地区洛浦县的均名为“白冬麦”的品种遗传距离最小, 为0.093; 来自阿克苏地区温宿县的“红冬麦”与来自喀什地区叶城县的白冬麦遗传距离最大, 为0.274。
第V类23个品种, 遗传距离在0.003~0.137之间, 包括红壳冬麦和白壳冬麦两种类型, 仅有2份白冬麦来自南疆喀什地区, 其余21份材料均来自北疆地区, 且有14份材料来自昌吉州, 表现为地域相近的材料聚在一起。其中来自乌鲁木齐市的“白冬麦”与阿勒泰地区布尔津县的晚熟冬麦之间的遗传距离最小, 为0.003; 来自昌吉州玛纳斯县的“红冬麦”与一份来自昌吉市的“白冬麦”之间的遗传距离最大, 为0.137。
第VI类18个品种, 遗传距离在0.003~0.111之间, 以白壳类型为主, 除了1个品种来自吐鲁番、2个品种来自和田地区外, 其余15个品种均来自南疆的阿克苏和巴州两个地区, 表现出明显的地域性。
第VII类25个品种, 遗传距离在0.003~0.125之间, 来自库尔勒市的买洛瓦西与来自和田市的早熟冬麦之间的遗传距离最小, 来自阿克苏地区新和县的梁山冬麦与喀什地区巴楚县的巴楚切利麦之间的遗传距离最大。此类包括白壳和红壳两种壳色类型, 分3个亚类, 第1亚类4个品种均来自巴州, 第2亚类9个品种中4个来自喀什地区、4个来自阿克苏地区、1个来自巴州; 第3亚类12个品种中有10份来自喀什地区, 也表现为地域相近的品种聚在一起。
第VIII类28个品种, 遗传距离在0.003~0.122之间, 包括红壳和白壳两种类型, 也分为3个亚类, 第1亚类9个品种中7个来自喀什地区; 第2亚类10个品种中有6个来自喀什地区, 其余4个品种分别来自塔城、阿勒泰、巴州和阿克苏地区; 第3亚类11个品种中6份来自喀什地区, 3份来自巴州, 2份来自阿克苏地区。
第IX类24个品种, 遗传距离在0.116~0.287之间, 来自昌吉州玛纳斯县的玛纳斯红冬麦与哈密地区哈密市的白芒冬麦之间的遗传距离最小, 来自喀什地区疏勒县的“白冬麦”与来自昌吉市的昌吉小红冬麦之间的遗传距离最大。分为3个亚类, 第1亚类12个品种中8个为红壳类型, 4个白壳类型, 其中8个品种来自吐鲁番, 3个品种来自与吐鲁番相邻的昌吉和哈密地区, 1个品种来自喀什地区。第2亚类7个品种中有2个来自北疆的塔城地区乌苏县, 5个均来自南疆的阿克苏、和田和巴州3个地区。第3亚类5个品种均来自南疆, 有4个品种来自巴州地区, 1个品种来自和田地区。
第X类最少, 仅有4个品种, 遗传距离在0.038~ 0.095之间, 3个来自和田地区, 1个来自哈密地区, 2个白壳类型聚在一起, 2个红壳类型聚在一起。
3 讨论
小麦地方品种是小麦育种的重要种质资源, 是改良品种的基础。地方品种是在自然经济农业时期, 在某地长期种植的“土著”品种, 由于各地生态条件千差万别, 造成地方品种具有不同的生态适应性[13,14]。新疆冬小麦地方品种多为强冬性, 具有耐寒、耐旱、耐盐碱、适应性强的特点, 但存在不耐肥、易倒伏、严重感锈病等缺陷, 逐渐被引进品种和自育品种所代替。育成品种(品系)往往都是丰产性、适应性、抗病性的结合体, 综合性状好, 是最便于利用的种质。在小麦育种中, 由于人们总是按照一定目标, 沿着一定方向选择, 使杂交育种亲本越来越集中到对当地条件最能适应, 综合性状最好, 配合力最佳的少数几个品种上, 导致育成品种遗传基础越来越狭窄[15]。如19世纪60年代, 新疆的小麦育种者利用具有抗寒、抗旱、耐盐碱特点的地方品种热衣木夏, 与引自丹麦的具有多花多实特性的冬小麦品种亥恩·亥德杂交育成新冬2号等品种及其衍生品种在南北疆大面积种植。本研究选用的54个新疆冬小麦育成品种中, 直接或间接以新冬2号为亲本选育成的就有新冬6号、新冬7号、新冬10号、新冬11号、新冬15号、奎冬4号6个品种, 这6个品种与其他42个育成品种聚在一类, 但新冬2号并没有与这6个品种聚在一类, 而是与新冬3号、新冬13号、巴冬1号等6个主要以新疆冬小麦地方品种与国外引进小麦品种为亲本育成的品种聚为一类(图3)。王立新等[16]对2005—2009年间547个国家冬小麦区域试验品系及134个国审品种分子标记遗传相似系数(GS)进行比较, 发现不同年度国家冬小麦区域试验GS>0.90的品种(系) 大多来自同一地区的育种单位。由此可见, 小麦育种中经常使用少数骨干亲本, 会导致小麦品种遗传多样性降低。李小军等[17]利用363对SSR标记对早洋麦、阿勃、欧柔和洛夫林10号等11份国外引种的骨干亲本材料及其衍生的33份代表性品种进行多样性分析, 发现衍生品种中约76.3%的等位变异类型均来自于国外亲本, 说明引种国外亲本对提高我国小麦品种的遗传多样性具有重要作用。因小麦地方品种多为人们根据品种表型特征如颖壳色、种皮色、芒色+熟性/地名等方式命名, 且随人员流动而传播, 因此多存在同物异名或同名异物现象, 需要通过田间种植表型观察结合分子标记方法来判断区分。王琨等[18]对长期种植于陕西关中不同生态种植区的6个同名地方品种蚂蚱麦进行农艺性状遗传变异分析, 认为不同来源的地方品种蚂蚱麦在形态和农艺性状表现上的差异不大, 但差异程度与地理位置有关。李正玲等[19]利用SSR标记对河南省15组名称相同的小麦地方品种共计155份材料进行组间和组内遗传多样性分析发现, 同名品种组内15组材料的平均遗传相似系数变化范围为0.66~0.93; 同名品种组间遗传相似系数分布范围为0.75~1.00, 认为小麦地方品种具有丰富的遗传多样性, 同名小麦地方品种间存在同名同质和同名异质的现象。本研究所选134个地方品种中, 名为“白冬麦”的有26个, 名为“红冬麦”的有17个, 名为“黑冬麦”的有9个, 根据品种间遗传距离和聚类结果, 证明同名品种间有差异, 品种间遗传差异与地理位置有关, 这与王琨等[14]的研究结果一致。
目前国内多以SSR标记开展小麦地方品种遗传多样性研究[20,21,22,23,24], 针对小麦地方品种材料仅见利用660K SNP标记进行研究的报道[25,26]。王宏飞等[22]利用42对SSR引物对75份新疆小麦地方品种进行遗传多样性分析, 共检测到317个等位变异, 等位变异变化范围为2~15个, 平均7.55个, PIC值变化范围为0.169~0.905, 平均0.696; 基因组的平均等位变异是D>B>A, 春小麦地方品种的遗传多样性高于冬小麦地方品种, 聚类结果反映了一定的地域特性。Zhou等[25]为了揭示中国小麦的传播历史、适应性进化和品种选择, 利用27,933个DArTseq标记和312,831个660K SNP标记分析来自中国10个农业生态区的717个中国小麦地方品种的遗传结构, 证明小麦首先是从中国西北部引进种植的, 环境压力和独立选择导致了中国小麦地方品种在种群水平上的差异, 群体遗传结构在很大程度上与中国小麦地方品种的地理分布相一致, 大多数具有亲缘关系的小麦地方品种来源地相近, 与各小麦种植区的小麦栽培历史一致; 在A、B、D三个基因组中, D基因组的基因多样性少于A和B基因组; 在单条染色体上, 中国小麦地方品种端粒的遗传变异远高于近着丝粒。陕西关中蚂蚱麦和山西平遥小白麦是我国北方小麦品种的原始骨干亲本, 白彦明等[26]利用小麦660K SNP芯片对陕西关中蚂蚱麦和山西平遥小白麦及其衍生品种(系)共计149份材料进行全基因组扫描, 发现来自黄淮冬麦区的4份蚂蚱麦与来自北部冬麦区的小白麦及其系选品种燕大1817的平均遗传相似系数GS为0.750, 高于蚂蚱麦和小白麦衍生系的GS (平均为0.619), 与本研究中育成品种的遗传多样性高于地方品种的结论一致。针对小麦地方品种与育成品种之间的遗传差异, 魏育明等[24]利用24个SSR标记对8份四川小麦地方品种和8份主栽品种的遗传多样性比较研究, 表明四川小麦地方品种群体内的遗传多样性(平均GS为0.812)低于主栽小麦品种群体内的遗传多样性(平均GS为0.670), 也与本研究的结论相似。推测原因, 可能是在早期较低的生产水平和环境条件下, 通过长期的人工和自然选择造成的地方品种等位基因趋同化。因此, 在育种过程中应充分利用外来种质资源, 实现外来种质中优异基因资源的导入和聚合, 拓宽当地小麦品种的遗传基础。
SNP标记作为第三代分子标记, 因其具有自动化程度高、通量大、速度快、易于建立标准化操作等优势, 已成为研究种质资源遗传多样性、遗传结构评估以及推断个体间祖先关系的有力工具[27,28]。国内目前利用SNP标记基因分型开展遗传多样性分析多见于以小麦育成品种为材料的报道[29,30,31]。刘易科等[29]利用高密度SNP芯片, 分别对国内不同麦区的229份冬小麦品种(系)进行遗传多样性研究, 发现长江中下游麦区小麦品种(系)的相似性系数最高(0.712), 黄淮南片麦区、黄淮北片麦区和北部麦区的小麦品种(系)的相似性系数相对较低(0.660~0.686), 可能与不同区域小麦育种单位实力以及引进小麦种质资源的遗传多样性有关。李珊珊等[30]利用小麦90K基因芯片对143份河北省小麦育成品种的基因分型数据分析其亲缘关系与遗传多样性表明, 河北省143份小麦育成品种的遗传相似系数主要分布于0.52~0.73之间(占89.23%), 低于曹廷杰等[31,32]对河南省2000—2013年审定的96个小麦品种遗传多样性研究的结果(主要分布在0.63~0.83之间, 占95.94%)。从以上结果可以看出, 遗传相似性高的小麦品种具有相似的地域性或来源于同一育种单位, 表明近年来各育种单位育种材料趋同, 且相同或相似的骨干亲本高频被应用, 导致小麦品种遗传多样性降低。
国外也有利用SNP标记对小麦品种资源开展遗传多样性研究的报道。Zahra等[33]利用152K SNP芯片评估了188个来自西亚和北非的面包小麦品种的遗传多样性, UPGMA聚类分析将基因型明确分为两大类, 一类包含177个品种, 另一类仅11个品种, 种质之间的相似系数在0.30~0.99之间, 平均值为0.64。Bhatta等[34]利用47,526个SNP标记对包括人工合成小麦和面包小麦种质的143份材料进行遗传多样性分析表明, SNP标记均匀分布在A、B、D三个基因组中; 群体结构分析根据小麦种质的类型和地理起源分为3个不同的小麦基因型群(日本人工合成小麦组, CIMMYT人工合成小麦组和面包小麦组); 遗传多样性比较表明, 人工合成小麦比面包小麦品系高33%, 认为人工合成小麦可用于拓宽现代小麦品种的遗传多样性。
依据本研究中55K芯片结果, 在分布于21条染色体上的53,063个SNP位点中有50,743个被检测到多态性, 多态性比率高达95.62%, 远高于刘易科等[29]、李珊珊等[30]、曹廷杰等[31,32]利用90K SNP获得的多态性; 并且本研究中SNP标记在A、B、D基因组间分布比较均匀, 分别为17,702个, 16,943个和16,098个, 而前人的90K芯片的SNP标记在B和D基因组间存在较明显的不均匀分布现象[29,30,31,32]。其原因可能由于小麦55K SNP芯片是以国内小麦材料为基础开发的, 并且所用的材料有普通小麦、四倍体小麦以及小麦野生近缘种材料, 因此SNP位点在各染色体上的分布更加均匀。由此可见, 小麦55K SNP芯片可能更加适宜于对国内小麦种质材料开展遗传多样性研究。
4 结论
利用我国自主研发, 覆盖小麦全基因组的55K SNP芯片, 对134个新疆冬小麦地方品种和54个新疆冬小麦育成品种进行全基因组扫描, 根据品种间的遗传距离将188个品种分为 10个类群, 育成品种集中聚在2个类群, 地方品种聚在另外8个类群。新疆冬小麦育成品种之间的遗传多样性略大于地方品种, 但育成品种与地方品种之间的遗传差异较大, 说明在现代小麦育种中, 地方品种可作为进一步拓宽育成品种遗传基础的育种材料。本研究证明小麦55K SNP标记能够揭示小麦品种资源的遗传差异以及品种间的亲缘关系, 可为小麦育种亲本选配提供重要理论依据。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.3969/j.issn.1004-3268.2005.09.001URL [本文引用: 1]

河南省是我国小麦主产区.建国前,河南省小麦面积400万hm2左右,单产不足600 kg/hm2,20世纪50年代总产在400万t上下.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1126/science.274.5287.536URLPMID:8928008 [本文引用: 2]
DOI:10.1007/s00122-018-3164-9URLPMID:30109392 [本文引用: 2]

KEY MESSAGE: A high-density genetic map constructed with a wheat 55 K SNP array was highly consistent with the physical map of this species and it facilitated the identification of a novel major QTL for productive tiller number. Productive tiller number (PTN) plays a key role in wheat grain yield. In this study, a recombinant inbred line population with 199 lines derived from a cross between '20828' and 'Chuannong16' was used to construct a high-density genetic map using wheat 55 K single nucleotide polymorphism (SNP) array. The constructed genetic map contains 12,109 SNP markers spanning 3021.04 cM across the 21 wheat chromosomes. The orders of the genetic and physical positions of these markers are generally in agreement, and they also match well with those based on the 660 K SNP array from which the one used in this study was derived. The ratios of SNPs located in each of the wheat deletion bins were similar among the wheat 9 K, 55 K, 90 K, 660 K and 820 K SNP arrays. Based on the constructed maps, a novel major quantitative trait locus QPtn.sau-4B for PTN was detected across multi-environments in a 0.55 cM interval on 4B and it explained 17.23-45.46% of the phenotypic variance. Twenty common genes in the physical interval between the flanking markers were identified on chromosome 4B of 'Chinese Spring' and wild emmer. These results indicate that wheat 55 K SNP array could be an ideal tool in primary mapping of target genes and the identification of QPtn.sau-4B laid a foundation for the following fine mapping and cloning work.
DOI:10.3389/fpls.2018.00333URLPMID:29599793 [本文引用: 1]

Maximum tiller number and productive tiller number are important traits for wheat grain yield, but research involving the temporal expression of tiller number at different quantitative trait loci (QTL) levels is limited. In the present study, a population set of 371 recombined inbred lines derived from a cross between Chuan-Nong18 and T1208 was used to construct a high-density genetic map using a Wheat55K SNP Array and to perform dynamic QTL analysis of the tiller number at four growth stages. A high-density genetic map containing 11,583 SNP markers and 59 SSR markers that spanned 4,513.95 cM and was distributed across 21 wheat chromosomes was constructed. A total of 28 single environmental QTL were identified in the recombined inbred lines population, and among these, seven QTL were stable and used for multi-environmental and dynamic analysis. These QTL were mapped to chromosomes 2D, 4A, 4D, 5A, 5D, and 7D, respectively. Each QTL explained 1.63-21.22% of the observed phenotypic variation, with an additive effect from -20.51 to 11.59. Dynamic analysis showed that cqTN-2D.2 can be detected at four growth stages of tillering, explaining 4.92-17.16% of the observed phenotypic variations and spanning 13.71 Mb (AX-109283238-AX-110544009: 82189047-95895626) according to the physical location of the flanking markers. The effects of the stable QTL were validated in the recombined inbred lines population, and the beneficial alleles could be utilized in future marker-assisted selection. Several candidate genes for MTN and PTN were predicted. The results provide a better understanding of the QTL selectively expressing the control of tiller number and will facilitate future map-based cloning. 9.17% SNP markers showed best hits to the Chinese Spring contigs. It was indicated that Wheat55K Array was efficient and valid to construct a high-density wheat genetic map.
DOI:10.1186/s12864-019-6005-6URLPMID:31395029 [本文引用: 1]

BACKGROUND: As one of the most important food crops in the world, increasing wheat (Triticum aestivum L.) yield is an urgent task for global food security under the continuous threat of stripe rust (caused by Puccinia striiformis f. sp. tritici) in many regions of the world. Molecular marker-assisted breeding is one of the most efficient ways to increase yield. Here, we identified loci associated to multi-environmental yield-related traits under stripe rust stress in 244 wheat accessions from Sichuan Province through genome-wide association study (GWAS) using 44,059 polymorphic markers from the 55 K single nucleotide polymorphism (SNP) chip. RESULTS: A total of 13 stable quantitative trait loci (QTLs) were found to be highly associating to yield-related traits, including 6 for spike length (SL), 3 for thousand-kernel weight (TKW), 2 for kernel weight per spike (KWPS), and 2 for both TKW and KWPS, in at least two test environments under stripe rust stress conditions. Of them, ten QTLs were overlapped or very close to the reported QTLs, three QTLs, QSL.sicau-1AL, QTKW.sicau-4AL, and QKWPS.sicau-4AL.1, were potentially novel through the physical location comparison with previous QTLs. Further, 21 candidate genes within three potentially novel QTLs were identified, they were mainly involved in the regulation of phytohormone, cell division and proliferation, meristem development, plant or organ development, and carbohydrate transport. CONCLUSIONS: QTLs and candidate genes detected in our study for yield-related traits under stripe rust stress will facilitate elucidating genetic basis of yield-related trait and could be used in marker-assisted selection in wheat yield breeding.
DOI:10.1186/s12870-019-1764-4URLPMID:30991940 [本文引用: 1]

BACKGROUND: Stripe rust (also called yellow rust) is a common and serious fungal disease of wheat (Triticum aestivum L.) caused by Puccinia striiformis f. sp. tritici. The narrow genetic basis of modern wheat cultivars and rapid evolution of the rust pathogen have been responsible for periodic and devastating epidemics of wheat rust diseases. In this study, we conducted a genome-wide association study with 44,059 single nucleotide polymorphism markers to identify loci associated with resistance to stripe rust in 244 Sichuan wheat accessions, including 79 landraces and 165 cultivars, in six environments. RESULTS: In all the field assessments, 24 accessions displayed stable high resistance to stripe rust. Significant correlations among environments were observed for both infection (IT) and disease severity (DS), and high heritability levels were found for both IT and DS. Using mixed linear models, 12 quantitative trait loci (QTLs) significantly associated with IT and/or DS were identified. Two QTLs were mapped on chromosomes 5AS and 5AL and were distant from previously identified stripe rust resistance genes or QTL regions, indicating that they may be novel resistance loci. CONCLUSIONS: Our results revealed that resistance alleles to stripe rust were accumulated in Sichuan wheat germplasm, implying direct or indirect selection for improved stripe rust resistance in elite wheat breeding programs. The identified stable QTLs or favorable alleles could be important chromosome regions in Sichuan wheat that controlled the resistance to stripe rust. These markers can be used molecular marker-assisted breeding of Sichuan wheat cultivars, and will be useful in the ongoing effort to develop new wheat cultivars with strong resistance to stripe rust.
DOI:10.1073/pnas.81.24.8014URLPMID:6096873 [本文引用: 1]

Spacer-length (sl) variation in ribosomal RNA gene clusters (rDNA) was surveyed in 502 individual barley plants, including samples from 50 accessions of cultivated barley, 25 accessions of its wild ancestor, and five generations of composite cross II (CCII), an experimental population of barley. In total, 17 rDNA sl phenotypes, made up of 15 different rDNA sl variants, were observed. The 15 rDNA sl variants comprise a complete ladder in which each variant differs in length from adjacent variants by approximately equal to 115 nucleotide pairs. Studies of four rDNA sl variants in an F2 population showed that these variants are located at two unlinked loci, Rrn1 and Rrn2, each with two codominant alleles. Using wheat-barley addition lines, we determined that Rrn1 and Rrn2 are located on chromosomes 6 and 7, respectively. The nonrandom distribution of sl variants between loci suggests that genetic exchange occurs much less frequently between than within the two loci, which demonstrates that Rrn1 and Rrn2 are useful as new genetic markers. Frequencies of rDNA sl phenotypes and variants were monitored over 54 generations in CCII. A phenotype that was originally infrequent in CCII ultimately became predominant, whereas the originally most frequent phenotype decreased drastically in frequency, and all other phenotypes originally present disappeared from the population. We conclude that the sl variants and/or associated loci are under selection in CCII.
DOI:10.1093/bioinformatics/bti282URLPMID:15705655 [本文引用: 1]

SUMMARY: PowerMarker delivers a data-driven, integrated analysis environment (IAE) for genetic data. The IAE integrates data management, analysis and visualization in a user-friendly graphical user interface. It accelerates the analysis lifecycle and enables users to maintain data integrity throughout the process. An ever-growing list of more than 50 different statistical analyses for genetic markers has been implemented in PowerMarker. AVAILABILITY: www.powermarker.net
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.tplants.2015.10.012URLPMID:26559599 [本文引用: 2]

Plant landraces represent heterogeneous, local adaptations of domesticated species, and thereby provide genetic resources that meet current and new challenges for farming in stressful environments. These local ecotypes can show variable phenology and low-to-moderate edible yield, but are often highly nutritious. The main contributions of landraces to plant breeding have been traits for more efficient nutrient uptake and utilization, as well as useful genes for adaptation to stressful environments such as water stress, salinity, and high temperatures. We propose that a systematic landrace evaluation may define patterns of diversity, which will facilitate identifying alleles for enhancing yield and abiotic stress adaptation, thus raising the productivity and stability of staple crops in vulnerable environments.
[本文引用: 1]
[本文引用: 1]
DOI:10.3724/SP.J.1006.2010.01490URL [本文引用: 1]

GS)和主要农艺性状,发现GS大于0.95的绝大多数品种(系)间主要农艺性状和表型高度相似,分子标记比对结果与田间比较结果吻合程度高达95%。依此确立了用分子标记筛查相似品种的方法体系,解决了无法从大量品种中查找相似品种(系)的问题。]]>
[本文引用: 1]
DOI:10.3724/SP.J.1006.2009.00778URL [本文引用: 1]

利用363对SSR标记分析了在我国小麦生产和育种中发挥了重要作用的11份国外引进品种和33份选育品种的遗传组成,旨在揭示国外种质对我国小麦品种改良的遗传贡献,指导种质资源引进和利用。国外种质包含了选育品种所发现等位变异的76.3%。与不同时期小麦品种等位基因多样性比较发现,国外种质的平均等位变异数最多(3.92),20世纪60年代(2.86)和70年代(3.01)基本一致,80年代有所升高(3.46)。品种间遗传距离比较与品种等位基因多样性结果相吻合。比较引进和选育品种在SSR位点的等位变异频率变化,发现至少在33个SSR位点,国外种质等位变异在我国小麦育种中被优先选择(该等位变异在引进和选育品种的分布频率均高于70%),其中一些位点已知与产量、生育期和抗病等性状密切相关。表明引进品种在以上基因组区域对我国小麦品种具有非常高的遗传贡献。]]>
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
华中农业大学硕士学位论文,
[本文引用: 1]
MS Thesis of Huazhong Agricultural University,
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
四川农业大学硕士学位论文,
[本文引用: 1]
MS Thesis of Sichuan Agricultural University,
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1111/pbi.12770URLPMID:28635103 [本文引用: 2]

Wheat was introduced to China approximately 4500 years ago, where it adapted over a span of time to various environments in agro-ecological growing zones. We investigated 717 Chinese and 14 Iranian/Turkish geographically diverse, locally adapted wheat landraces with 27 933 DArTseq (for 717 landraces) and 312 831 Wheat660K (for a subset of 285 landraces) markers. This study highlights the adaptive evolutionary history of wheat cultivation in China. Environmental stresses and independent selection efforts have resulted in considerable genome-wide divergence at the population level in Chinese wheat landraces. In total, 148 regions of the wheat genome show signs of selection in at least one geographic area. Our data show adaptive events across geographic areas, from the xeric northwest to the mesic south, along and among homoeologous chromosomes, with fewer variations in the D genome than in the A and B genomes. Multiple variations in interdependent functional genes such as regulatory and metabolic genes controlling germination and flowering time were characterized, showing clear allelic frequency changes corresponding to the dispersion of wheat in China. Population structure and selection data reveal that Chinese wheat spread from the northwestern Caspian Sea region to South China, adapting during its agricultural trajectory to increasingly mesic and warm climatic areas.
DOI:10.3724/SP.J.1006.2019.91012URL [本文引用: 2]

陕西关中蚂蚱麦和山西平遥小白麦是我国北方小麦品种的原始骨干亲本, 解析蚂蚱麦和小白麦及其衍生系的遗传多样性对于小麦品种改良具有重要的参考意义。本研究利用小麦660K SNP芯片对蚂蚱麦、小白麦及其衍生品种(系)进行全基因组扫描, 分析其遗传多样性。结果表明, 小麦3个基因组的多态性SNP标记数为B>A>D, 第4同源群的多态性标记数最少, 149份供试材料基因多样性(H)范围为0.095~0.500, 平均值为0.336; 核苷酸多样性指数(π)范围为0.272~0.435, 平均值为0.340; 而遗传相似系数(GS)变幅为0.335~0.997, 平均值达0.619, 表明蚂蚱麦和小白麦衍生系的遗传多样性较低。聚类分析表明蚂蚱麦和小白麦紧密地聚在亚群I, 其衍生品种(系)分为5个亚群, 其中2000年以前以蚂蚱麦或小白麦的单一衍生系为主, 分在亚群I、II、III, 2000年以后多数品种同时拥有蚂蚱麦和小白麦血缘, 分在亚群IV、V, 遗传多样性较高, 且与大面积推广品种聚为一类。因此, 应加强优异基因资源导入, 拓宽小麦品种的遗传基础, 最终提高育种水平。
[本文引用: 2]
DOI:10.1111/pbi.12183URL [本文引用: 1]

High-density single nucleotide polymorphism (SNP) genotyping arrays are a powerful tool for studying genomic patterns of diversity, inferring ancestral relationships between individuals in populations and studying marker-trait associations in mapping experiments. We developed a genotyping array including about 90 000 gene-associated SNPs and used it to characterize genetic variation in allohexaploid and allotetraploid wheat populations. The array includes a significant fraction of common genome-wide distributed SNPs that are represented in populations of diverse geographical origin. We used density-based spatial clustering algorithms to enable high-throughput genotype calling in complex data sets obtained for polyploid wheat. We show that these model-free clustering algorithms provide accurate genotype calling in the presence of multiple clusters including clusters with low signal intensity resulting from significant sequence divergence at the target SNP site or gene deletions. Assays that detect low-intensity clusters can provide insight into the distribution of presence-absence variation (PAV) in wheat populations. A total of 46 977 SNPs from the wheat 90K array were genetically mapped using a combination of eight mapping populations. The developed array and cluster identification algorithms provide an opportunity to infer detailed haplotype structure in polyploid wheat and will serve as an invaluable resource for diversity studies and investigating the genetic basis of trait variation in wheat.
DOI:10.1186/1471-2164-11-702URLPMID:21156062 [本文引用: 1]

BACKGROUND: A genome-wide assessment of nucleotide diversity in a polyploid species must minimize the inclusion of homoeologous sequences into diversity estimates and reliably allocate individual haplotypes into their respective genomes. The same requirements complicate the development and deployment of single nucleotide polymorphism (SNP) markers in polyploid species. We report here a strategy that satisfies these requirements and deploy it in the sequencing of genes in cultivated hexaploid wheat (Triticum aestivum, genomes AABBDD) and wild tetraploid wheat (Triticum turgidum ssp. dicoccoides, genomes AABB) from the putative site of wheat domestication in Turkey. Data are used to assess the distribution of diversity among and within wheat genomes and to develop a panel of SNP markers for polyploid wheat. RESULTS: Nucleotide diversity was estimated in 2114 wheat genes and was similar between the A and B genomes and reduced in the D genome. Within a genome, diversity was diminished on some chromosomes. Low diversity was always accompanied by an excess of rare alleles. A total of 5,471 SNPs was discovered in 1791 wheat genes. Totals of 1,271, 1,218, and 2,203 SNPs were discovered in 488, 463, and 641 genes of wheat putative diploid ancestors, T. urartu, Aegilops speltoides, and Ae. tauschii, respectively. A public database containing genome-specific primers, SNPs, and other information was constructed. A total of 987 genes with nucleotide diversity estimated in one or more of the wheat genomes was placed on an Ae. tauschii genetic map, and the map was superimposed on wheat deletion-bin maps. The agreement between the maps was assessed. CONCLUSIONS: In a young polyploid, exemplified by T. aestivum, ancestral species are the primary source of genetic diversity. Low effective recombination due to self-pollination and a genetic mechanism precluding homoeologous chromosome pairing during polyploid meiosis can lead to the loss of diversity from large chromosomal regions. The net effect of these factors in T. aestivum is large variation in diversity among genomes and chromosomes, which impacts the development of SNP markers and their practical utility. Accumulation of new mutations in older polyploid species, such as wild emmer, results in increased diversity and its more uniform distribution across the genome.
[本文引用: 4]
[本文引用: 4]
[本文引用: 4]
[本文引用: 4]
DOI:10.3724/SP.J.1006.2015.00197URL [本文引用: 4]

为了解河南省近年小麦品种的遗传基础,利用Illumina 90k iSelect SNP标记技术对豫麦34及该省2000—2013年审定的小麦品种共96个进行全基因组扫描,分析了其遗传多样性和遗传基础。结果表明,在所有SNP位点中,多态性比率为47.39% (38 661/81 587),多态性标记在基因组间分布呈现B>A>D。96个品种亲缘关系较近,两两遗传相似系数的平均值为0.719,变幅为0.552~0.998,且94.3%的品种间遗传相似系数在0.652~0.812之间;按UPGMA法将96个品种划分为7个类群。综合SNP和系谱分析,近10年河南省审定的96个小麦品种遗传多样性不够丰富,多数品种亲缘关系较近,在育种中迫切需要引入新的种质资源,拓宽遗传背景。
[本文引用: 4]
[本文引用: 3]
[本文引用: 3]
DOI:10.1007/s10722-019-00836-zURL [本文引用: 1]
DOI:10.1007/s13353-019-00514-xURLPMID:31414379 [本文引用: 1]

Recurrent selection and intercrossing between best of the best parents in each generation of breeding cycle resulted in a narrower genetic diversity in elite wheat (Triticum aestivum L.) germplasm. Therefore, we investigated diverse source of 143 synthetic and bread wheat accessions for identifying potentially rich genetic resources for improving the genetic diversity in wheat. This study identified 47,526 genotyping-by-sequencing-derived SNP markers that were nearly evenly distributed across three genomes of wheat. The population structure analysis identified three distinct clusters (Japan synthetics, CIMMYT synthetics, and bread wheat) of wheat genotypes on the basis of type and geographical origin of wheat accessions. Population differentiation using analysis of molecular variance indicated 21% of the total genetic variance among subgroups and the remainder within subgroups. This study also identified that the Japan synthetic group was the most divergent group compared with other subgroups. The genetic diversity comparisons between synthetic and bread wheat lines showed that the gene diversity of synthetic wheat was 33% higher than bread wheat accessions, indicating the potential use of these lines for broadening the genetic diversity of modern wheat cultivars. The results from this study will be helpful in further understanding genomic features of wheat and facilitate their use in wheat breeding programs.
