
 , 童金蓉1, 黄振华1, 刘静1, 刘海泉1,3,4, 潘迎捷1,3,4, 赵勇1,3,4
, 童金蓉1, 黄振华1, 刘静1, 刘海泉1,3,4, 潘迎捷1,3,4, 赵勇1,3,4

1. 上海海洋大学 食品学院,上海 201306;
2. 上海海洋大学 水产与生命学院,上海 201306;
3. 农业农村部水产品贮藏保鲜质量安全风险评估实验室 (上海),上海 201306;
4. 上海水产品加工及贮藏工程技术研究中心,上海 201306
收稿日期:2020-08-11;接收日期:2020-10-21;网络出版时间:2020-11-09
基金项目:国家博士后创新人才支持计划(No. BX20190194),国家自然科学基金(Nos. 32001800,31972188),上海市“超级博士后”激励计划(No. 2019348),中国博士后科学基金(No. 2019M661469),上海市教育委员会科研创新计划(No. 2017-01-07-00-10-E00056)
摘要:食源性致病菌对人类健康与公众安全造成了极大的危害,形成生物被膜加剧了它们的致病与耐药风险。酶具有高度专一性,可靶向作用于生物被膜中的特殊物质,从而清除食源性致病菌的生物被膜,具有重要的科研价值和广泛的应用前景。因此,文中系统地综述了相关酶制剂清除食源性致病菌生物被膜的研究进展。根据酶制剂的不同作用靶点,着重介绍了群体感应抑制酶、环二鸟苷酸代谢酶、胞外基质水解酶等酶制剂的研究现状。文中还针对抗生物被膜酶制剂的未来研究方向进行了展望,旨在为食源性致病菌生物被膜的有效控制提供新的技术与策略。
关键词:酶制剂食源性致病菌生物被膜靶向清除
Enzyme-based targeted disintegration of biofilms formed by food-borne pathogens: a review
Qian Wu1, Zhaohuan Zhang1,2

 , Jinrong Tong1, Zhenhua Huang1, Jing Liu1, Haiquan Liu1,3,4, Yingjie Pan1,3,4, Yong Zhao1,3,4
, Jinrong Tong1, Zhenhua Huang1, Jing Liu1, Haiquan Liu1,3,4, Yingjie Pan1,3,4, Yong Zhao1,3,4

1. College of Food Science and Technology, Shanghai Ocean University, Shanghai 201306, China;
2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China;
3. Laboratory of Quality & Safety Risk Assessment for Aquatic Product on Storage and Preservation (Shanghai), Ministry of Agriculture and Rural Affairs, Shanghai 201306, China;
4. Shanghai Engineering Research Centre of Aquatic-Product Processing & Preservation, Shanghai 201306, China
Received: August 11, 2020; Accepted: October 21, 2020; Published: November 9, 2020
Supported by: National Postdoctoral Program for Innovative Talents of China (No. BX20190194), National Natural Science Foundation of China (Nos. 32001800, 31972188), Shanghai Post-doctoral Excellence Program, China (No. 2019348), China Postdoctoral Science Foundation (No. 2019M661469), Innovation Program of Shanghai Municipal Education Commission, China (No. 2017-01-07-00-10-E00056)
Corresponding author: Zhaohuan Zhang. Tel: +86-21-61900503; E-mail: gongziwuhen@126.com;
Yong Zhao. Tel: +86-21-61900768; E-mail: yzhao@shou.edu.cn.
Abstract: Food-borne pathogens pose great risks to human health and public safety, and the formation of biofilm exacerbates their pathogenicity and antimicrobial resistance. Enzymes can target special substances in the biofilm to disintegrate the biofilm of food-borne pathogens, which has great potential for applications. This review summarized the progress of using enzymes to disintegrate the biofilms of food-borne pathogens, highlighting quorum-quenching enzymes, C-di-GMP metabolic enzymes, as well as extracellular matrix hydrolases. Finally, challenges and perspectives on developing enzymes into effective products for disintegrating the biofilms of food-borne pathogens were discussed.
Keywords: enzymesfoodborne pathogensbiofilmtargeted disintegration
食源性致病菌导致的食源性疾病是全世界共同关注的焦点、难点问题。据世界卫生组织(World Health Organization,WHO) 统计,全球每年约有42万人因感染食源性致病菌而死亡[1]。食源性致病菌能够在食品基质及相关加工器械表面形成生物被膜,这种特殊的结构可以保护病原体免受抗生素、洗涤剂、高温高压等不利外界条件的侵害[2]。生物被膜是由细菌细胞黏附于接触物体表面,通过分泌多糖、蛋白、脂质等胞外聚合物而形成的多细胞聚集体[3-4]。研究表明,当细菌存在于生物被膜中时,它们对抗生素的耐药性会提高10–1 000倍[5-6],笔者课题组研究表明,生物被膜可为细菌提供耐药基因突变和水平基因转移的机会,加剧食源性致病菌的耐药风险[7-8]。此外,生物被膜的形成还容易引起食品腐败、食品相关设备腐蚀、交叉污染等严重的食品安全问题,造成巨大的经济损失[9]。
传统食品工业中,常用化学试剂或物理杀菌等方式来减轻生物被膜的危害[10],但是残留的化学试剂容易导致环境污染[11],而一些物理杀菌方式有可能破坏食品的质构、改变食品的风味[12]。酶与底物的结合具有高度特异性,能够靶向作用于生物被膜的胞外基质,降解生物被膜中的群体感应信号分子,阻断细胞与细胞之间的交流,从而加速生物被膜的瓦解[13]。酶制剂具有专一、高效、无污染等优势,已经广泛应用于食品工业之中[14]。图 1总结了酶制剂清除食源性致病菌生物被膜的作用机制,首先酶通过渗透作用进入病原体的生物被膜,然后特异性地识别特殊物质,从根本上清除食源性致病菌的生物被膜。
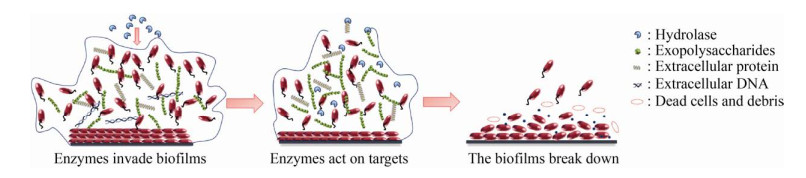 |
| 图 1 酶制剂清除食源性致病菌生物被膜的动态过程 Fig. 1 Process of enzyme-targeted disintegration of biofilm of food-borne pathogens. |
| 图选项 |
因此,本文针对食源性致病菌生物被膜的危害,结合笔者课题组的相关研究,综述了应用酶制剂清除食源性致病菌生物被膜的研究进展,并根据酶制剂作用靶点及机制的不同,系统地将其分类为群体感应抑制酶、环二鸟苷酸代谢酶、胞外基质水解酶等,详细地阐述了这些酶的来源、分类、作用靶点及应用,并对其未来研究方向进行了展望。本文旨在为食源性致病菌生物被膜的有效控制提供新的思路,为抗生物被膜酶制剂的进一步开发与应用奠定理论基础。
1 食源性致病菌生物被膜的危害生物被膜能够导致食源性致病菌产生耐药性、增强致病菌对宿主细胞的侵袭力、造成食品工业中交叉污染等。首先,生物被膜的特殊微观结构和代谢状态,是细菌耐药性形成的因素之一[15]。Hall等[6]发现假单胞菌携带胞外多糖合成基因座(psl,polysaccharide synthesis locus),能够合成Psl胞外多糖,在该菌生物被膜形成阶段对多粘菌素B、环丙沙星等产生耐药性[13]。笔者课题组研究表明,副溶血性弧菌Vibrio parahaemolyticus和单核细胞增生李斯特菌Listeria monocytogenes形成混合生物被膜时,耐药性发生显著变化[8]。其次,生物被膜能增加致病菌毒性,提升其对宿主细胞的侵袭力[16]。例如,生物被膜会促使金黄色葡萄球菌Staphylococcus aureus释放大量肠毒素,导致食物中毒[17]。被膜态的霍乱弧菌Vibrio cholerae感染细胞时,毒力基因表达量显著提升[18]。第三,食源性致病菌可在食品加工各个环节形成生物被膜,导致交叉污染[9]。例如,金黄色葡萄球菌、假单胞菌Pseudomonas等会粘附于乳品生产线、运输管道等表面形成生物被膜,导致乳品的交叉污染和食品安全问题[19]。笔者课题组研究表明,副溶血性弧菌易在食品加工材料[20-21]及水产品[22]表面形成生物被膜,并能与霍乱弧菌[23]、单核细胞增生李斯特菌[8]等形成混合生物被膜,引起食品工业中的交叉污染问题。
2 用于清除生物被膜的酶制剂针对以上食源性致病菌生物被膜的严重危害,研究人员开发了不同种类的抗生物被膜酶制剂,为生物被膜的有效瓦解提供了新的技术和策略[14]。本文进一步对基于不同作用靶点的抗生物被膜酶制剂进行总结,其具体的作用靶点如图 2所示,这些酶制剂通过抑制或水解致病菌生物被膜形成过程中的必需物质,达到清除生物被膜的目的。其中包括群体感应抑制酶、环二鸟苷酸代谢酶和胞外基质水解酶等。
 |
| 图 2 不同靶点清除食源性致病菌生物被膜的酶制剂 Fig. 2 Categories of enzymes that are able to targeted-disintegrate the biofilm of food-borne pathogens. (A) Enzymes act at quorum sensing signaling molecules. (B) Enzymes act at C-di-GMP. (C) Hydrolases act at exopolysaccharides. (D) Hydrolases act at extracellular proteins. (E) Hydrolases act at extracellular DNA. |
| 图选项 |
2.1 群体感应抑制酶群体感应(Quorum sensing,QS) 是细菌之间相互交流的一种方式,大部分微生物可分泌信号分子调控细菌的行为,如形成生物被膜、产生毒素、产生抗菌肽等。群体感应信号分子可分为3类:一是N-乙酰高丝氨酸内酯(N-acyl-homoserine lactones,AHLs);二是自诱导肽(Autoinducing peptide,AIP);三是自诱导物-2 (Autoinducer-2,AI-2)[24]。AHLs是一类在革兰氏阴性菌群体感应系统中最典型的信号分子,在细菌生物被膜的形成中起着关键作用[25]。因此,靶向作用于群体感应信号分子的酶制剂能有效地清除食源性致病菌生物被膜。
群体感应抑制酶(Quorum quenching enzyme,QQ酶) 的作用机制是通过降解群体感应信号分子,达到抑制生物被膜形成的目的[26]。表 1总结了QQ酶的分离来源、致病菌种类及靶向作用位点。目前,大多数QQ酶的研究都以AHLs为作用位点,可以从芽孢杆菌、假单胞菌、拟杆菌等多种来源获得。根据酶制剂的特性,可系统地将降解AHLs的酶分为3类:AHLs-内酯酶、AHLs-酰化酶和AHLs-氧化还原酶。
表 1 群体感应抑制酶的种类、来源及作用靶点Table 1 The sources, target bacteria, and target quorum-sensing (QS) molecules of quorum-quenching (QQ) enzymes
| Quorum-quenching enzymes | Source | Target pathogenic bacteria | References |
| AHLs-lactonases | |||
| ???AiiA | Bacillus spp. strain 240B1 | Erwinia carotovora | [27] |
| ???AiiAB546 | Bacillus spp. strain B546 | Aeromonas hydrophila | [28] |
| ???AiiM | Microbacterium testaceum strain StLB037 | Pectobacterium carotovorum | [29] |
| ???AidH | Ochrobactrum spp. strain T63 | Pseudomonas fluorescens | [30] |
| ???DlhR and QsdR1 | Rhizobium spp. strain NGR234 | Pseudomona aeruginosa | [31] |
| Chromobacterium violaceum | |||
| Agrobacterium tumefaciens | |||
| ???BpiB01 | Nitrobacter spp. strain Nb-311A | Pseudomona aeruginosa | [32] |
| ???BpiB04 | Pseudomonas fluorescens | Pseudomona aeruginosa | [32] |
| AHLs-acylase | |||
| ???AiiD | Variovorax paradoxus VAI-C | Pseudomona aeruginosa | [33] |
| ???Aac | Shewanella spp. strain MIB015 | Vibrio anguillarum | [34] |
| ???Aac | Ralstonia solanacearum strain GMI1000 | Chromobacterium violaceum | [35] |
| ???HacB (PA0305) | Pseudomonas aeruginosa strain PAO1 | Pseudomonas aeruginosa | [36] |
| ???PvdQ (PA2385) | Pseudomonas aeruginosa strain PAO1 | Pseudomonas aeruginosa | [37] |
| Oxidoreductases | |||
| ???BpiB09 (Short-chain dehydrogenase/reductase [SDR]) | Acidobacterium spp. strain MP5ACTX8 | Pseudomonas aeruginosa | [38] |
| ???Reductase | Burkholderia sp. GG4; Klebsiella sp. strain Se14; Acinetobacter spp. strain GG2 | Pseudomonas aeruginosa; Erwinia carotovora | [39] |
表选项
2.1.1 AHLs-内脂酶AiiA酶是第一个被分离获得的AHLs-内脂酶,来源于芽孢杆菌240B1,通过水解方式裂解AHLs中的高丝氨酸内酯环,降低QS分子的有效性,对胡萝卜软腐欧文氏菌Erwinia carotovora的生物被膜有明显的抑制效果[27]。Chen等[28]从芽孢杆菌B546中也分离获得了AiiA酶,将其命名为AiiAB546,可有效抑制嗜水气单胞菌Aeromonas hydrophila的生物被膜。Wang等[29]在砖红色微杆菌StLB037 Microbacterium testaceum的染色体上发现了一段编码AHLs-内脂酶的基因(aiiM),通过重组表达获得了AiiM酶,可用于胡萝卜软腐果胶杆菌Pectobacterium carotovorum生物被膜的控制。
目前,在根瘤菌Rhizobium spp.、硝化杆菌Bacterium nitrobacter、荧光假单胞菌Pseudomonas fluorescens等细菌中也发现了AHLs-内脂酶。例如,根瘤菌NGR234产生的AHLs-内脂酶DlhR和QsdR1,可通过抑制群体感应来控制铜绿假单胞菌Pseudomonas fluorescens、紫色色杆菌Chromobacterium violaceum和根癌农杆菌Agrobacterium tumefaciens的生物被膜[31]。研究人员分别从硝化杆菌Nb-311A[32]和荧光假单胞菌[32]中分离获得了BpiB01和BpiB04,能够降解AHLs分子并抑制铜绿假单胞菌生物被膜的形成。
2.1.2 AHLs-酰化酶AHLs-酰化酶属于NTN-水解酶超家族,最早发现于争论贪噬菌VAI-C Variovorax paradoxus的生物被膜中,能水解AHLs酰基链和高丝氨酸部分之间的酰胺键[33]。Wahjud等[36]从铜绿假单胞菌中获得的HacB酶,对铜绿假单胞菌的长链AHLs分解具有催化活性,能抑制该菌生物被膜的形成。Sio等[37]从铜绿假单胞菌PAO1中获得的PvdQ酶,能降解该菌中含有11–14个碳链的AHLs,降低细胞毒性。
2.1.3 AHLs-氧化还原酶AHLs-氧化还原酶能够催化AHLs酰基侧链的氧化或还原[40-41]。目前,已在伯克霍尔德氏菌属Burkholderia spp.、克雷伯氏菌属Klebsiella spp.、不动杆菌属Acinetobacter spp.等细菌中发现AHLs-氧化还原酶。例如,Bijtenhoorn等[38]从酸杆菌属MP5ACTX8 Acidobacterium spp.中分离获得的BpiB09短链脱氢酶,该酶在铜绿假单胞菌PAO1中的表达,导致了铜绿假单胞菌运动能力降低、生物被膜形成量减少。
2.2 基于环二鸟苷酸的生物被膜代谢酶环二鸟苷酸(C-di-GMP) 是著名的细菌第二信使,存在于所有细菌之中[42],具有多种调节功能,如胞外聚合物(EPS) 生物合成、生物被膜的形成、参与细胞的运动等。研究表明C-di-GMP水平升高可使生物被膜形成能力显著增强[43],参与调节C-di-GMP浓度的代谢酶主要分为两类:二鸟苷酸环化酶(Diguanylate cyclases,DGCs) 和磷酸二酯酶(Phosphodiesterases,PDEs)。表 2总结了具有代表性的C-di-GMP代谢酶,合理利用DGCs和PDEs可加速C-di-GMP的降解,从而清除食源性致病菌的生物被膜。
表 2 环二鸟苷酸代谢酶Table 2 C-di-GMP metabolic enzymes
| The type of enzyme | Name | Source | Function | Domain | References |
| Diguanylate cyclase | PleD | Caulobacter crescentus | C-di-GMP synthesis | GGDEF domain | [44] |
| WspR | Pseudomonas aeruginosa | C-di-GMP synthesis; Biofilm formation and persistence | GGDEF domain | [45] | |
| DgcA, DgcB and DgcC | Listeria monocytogenes | Biofilm formation | GGDEF domain | [46] | |
| Phosphodiesterase | YkuI | Bacillus subtilis | C-di-GMP hydrolysis | EAL domain | [47] |
| BlrP1 | Klebsiella pneumoniae | C-di-GMP hydrolysis was regulated by light | EAL domain | [48] | |
| Bd1817 | Vermiculite isolated bacteria | Unknown | HD-GYP domain | [49] | |
| PdeB, PdeC and PdeD | Listeria monocytogenes | C-di-GMP hydrolysis | EAL domain | [46] |
表选项
在大多数细菌中,DGCs参与C-di-GMP的催化合成,而PDEs则负责C-di-GMP的催化降解。DGCs带有一个保守GGDEF (Gly-Gly-Asp-Glu-Phe) 结构域[42],能够催化两个GTP分子合成C-di-GMP。PDEs具有保守EAL (Glu-Ala-Leu) 或HD-GYP (His-Asp-Gly-Tyr-Pro) 结构域,可将C-di-GMP水解为线性的二聚鸟苷酸(pGpG),最后被寡核糖核酸酶分解为两个GMP分子。目前,尚未有研究将以上两种C-di-GMP代谢酶应用于致病菌生物被膜的控制,未来可根据这两种酶的生物学特性,催化加速C-di-GMP的合成与降解,达到清除生物被膜的目的。
2.3 基于胞外基质的生物被膜水解酶胞外基质是生物被膜的主要成分,能为生物被膜中的致病菌提供保护,加强其对宿主免疫系统的防御和对抗生素的抵抗作用[50-51]。笔者课题组研究表明,胞外聚合物在副溶血性弧菌生物被膜形成过程中起决定性作用[52-53]。胞外基质水解酶的作用机制是利用酶的专一性,靶向作用于胞外基质成分,从而达到清除食源性致病菌生物被膜的目的。表 3中总结了各种胞外基质水解酶的来源与作用靶点。胞外基质水解酶主要分为3类:胞外多糖水解酶、胞外蛋白水解酶和胞外DNA水解酶。
表 3 胞外基质水解酶的种类、来源和作用靶点Table 3 The categories, sources and targets of extracellular matrix hydrolases
| Extracellular matrix | Category | Source | Target/Matrix | References |
| Exopolysaccharides | Alginate lyase | Pseudomonas aeruginosa | β-glycoside bond that links a brown alginate polymer | [54] |
| PslGh | Pseudomonas aeruginosa | Psl exopolysaccharides | [55] | |
| PelAh | Pseudomonas aeruginosa | Pel exopolysaccharides | [55] | |
| Lysozyme | Microbial, animal and plant extracts | Peptidoglycan layer of bacterial cell wall | [56] | |
| α-Amylase | Bacillus subtilis | α-1, 4 glycosidic linkage | [57] | |
| Dispersing B | Periodontal actinomycetes | β-1, 6-N-acetyl-d-glucosamine | [58] | |
| Extracellular protein | Bacillus subtilis protease | Bacillus | Peptide bonds in protein structures | [59] |
| Lysostaphin | Staphylococcus aureus | Pentapeptide bond of peptidoglycan layer | [60] | |
| Bacteriophage lysin | Bacteriophage Vb-SepiS-phiIPLA7 | Bacterial cell walls based on peptidoglycan | [61] | |
| Extracellular DNA | DNase | Widespread in living organisms | The phosphodiester linkage of DNA | [62] |
表选项
2.3.1 基于胞外多糖的生物被膜水解酶胞外多糖是构筑食源性致病菌生物被膜的核心骨架,能帮助生物被膜中的细菌进行粘附,促进其在食品器械表面的定植。利用胞外多糖水解酶针对性地水解胞外多糖,有助于破坏细菌的附着,减少生物被膜的形成量。在铜绿假单胞菌的生物被膜基质中存在3种活性胞外多糖:褐藻胶、Psl和Pel (一种由N-乙酰-d-氨基葡萄糖和N-乙酰基-d-半乳糖胺组成的阳离子胞外多糖)[63]。研究表明,褐藻胶裂解酶(AlsL) 破坏生物被膜中褐藻胶聚合物的作用位点是β-糖苷键,可加速假单胞菌生物被膜的催化降解,从非生物表面剥离生物被膜[64-65],并具有增强人体免疫细胞杀菌效果的功能[66]。Baker等[55]研究发现,Pel和Psl生物合成操纵子中分别存在一段可编码糖苷水解酶PelAh和PslGh的基因序列,这两种水解酶能够分别靶向降解生物被膜中的胞外多糖Pel和Psl,抑制生物被膜的形成。此外,Asker等[67]将糖苷水解酶PslGh固定在玻璃、PDMS等材料表面,构建了一种铜绿假单胞菌生物被膜完全不能生长的新型生物材料。
2.3.2 基于胞外蛋白的生物被膜水解酶胞外蛋白作为生物被膜胞外基质的主要成分之一,在食源性致病菌生物被膜的结构和功能中发挥重要作用[68]。蛋白水解酶能够通过水解氨基酸残基之间的特定肽键降解胞外蛋白,从而达到清除生物被膜的效果。枯草芽孢杆菌Bacillus subtilis蛋白酶[58]是一种能够攻击肽键的非特异性丝氨酸蛋白酶,能破坏假交替单胞菌Pseudoalteromonas的粘附性胞外蛋白,使生物被膜的结构分离。此外,溶葡萄球菌酶(Lst)[69]是一种应用广泛的锌依赖性金属蛋白酶,可水解食源性致病菌肽聚糖层的五聚肽键,达到降解胞外蛋白的目的。
2.3.3 基于胞外DNA的生物被膜水解酶胞外DNA (eDNA) 是食源性致病菌生物被膜形成的关键元素[70],在单核细胞增生李斯特菌[71]、铜绿假单胞菌[72]和金黄色葡萄球菌[73]生物被膜的粘附、构建和维护中起着重要作用,因此降解eDNA也是清除生物被膜的一种有效的方法。商用的脱氧核糖核酸酶(DNase Ⅰ) 通常用于破坏DNA的磷酸二酯键,在DNase Ⅰ处理革兰氏阴性菌和革兰氏阳性菌的实验中发现[74],5 μg/mL的DNase Ⅰ可以使细菌的生物被膜量减少约40%。此外,DNase Ⅰ还可与其他酶联合使用,Karygianni等[75]研究发现DNase Ⅰ和蛋白酶联合处理能破坏口腔生物被膜组成及结构的完整性。
2.4 其他类型的生物被膜水解酶2.4.1 氧化还原酶氧化还原酶是利用H2O2抑制细菌的生长繁殖,从而减少生物被膜的形成。葡萄糖氧化酶(Gox)能催化葡萄糖氧化反应,产生具有抗菌活性的H2O2和葡萄糖内酯,降低细菌生存环境的pH值,使大肠杆菌、金黄色葡萄球菌、单核细胞增生李斯特菌等食源性致病菌的生长受到抑制[76]。纤维二糖脱氢酶(Cdh) 是另一种氧化还原酶,能产生有细胞毒性的H2O2,抑制细菌细胞及生物被膜的生长[77]。
2.4.2 脂肪水解酶脂质是许多细菌生物被膜中常见的基质成分,主要参与生物被膜表面的相互作用、附着和维持。从大蜡螟中分离出的脂肪酶对结核分枝杆菌Mycobacterium tuberculosis H37Rv具有杀灭作用,从牛胰腺中产生的脂肪酶可降解胞外基质,防止致病菌在海洋环境中形成生物被膜[78]。此外,有研究表明[79],海洋杆菌属PUMB02 Oceanobacillus spp.衍生的脂肪酶能够抑制食品加工环境中生物被膜的形成。
3 酶制剂与其他技术联合作用于生物被膜的研究酶的专一性、高效性使其成为清除食源性致病菌生物被膜的绝佳手段,在实际应用过程中,许多研究者常将酶制剂与其他技术联合使用,包括多种酶制剂联合使用、酶制剂与化学试剂联合使用、酶制剂与物理杀菌方式联合使用等。这种联合作用的方式,拓展了酶制剂的多元应用功效,不仅能最大程度地清除生物被膜,还能有效地杀灭残留的致病菌,为食品质量安全控制提供了可靠的技术支撑。
3.1 多种酶制剂联合使用不同酶制剂的作用靶点不同,如QQ酶抑制QS分子的积累、C-di-GMP代谢酶催化调节C-di-GMP浓度、胞外基质水解酶水解胞外基质等,均能达到清除食源性致病菌生物被膜的目的。若将多种酶制剂联合使用,则能提高其清除细菌生物被膜的效率。Nguyen等[80]研究了DNase Ⅰ和蛋白酶K对单核细胞增生李斯特菌生物被膜的清除作用,当使用100 μg/mL的DNase Ⅰ处理72 h的生物被膜时,生物被膜的残留量为25%。若将DNase Ⅰ与蛋白酶K联合使用,则能完全抑制生物被膜的形成。此外,Johansen等[81]研究表明,氧化还原酶与多糖水解酶的组合可以清除生物被膜并产生杀菌活性。
3.2 酶制剂与化学杀菌技术联合使用常规洗涤剂难以去除食品工业设备接口、缝隙等地方的生物被膜,若将酶制剂和洗涤剂联合使用,则可弥补传统洗涤剂的不足,提高洗涤剂的清洗效率。Walker等[82]研究表明,通过酶制剂的预处理,有助于洗涤剂更好地清除生产线中生物被膜的污染。酶制剂的存在还能提高化学试剂的杀菌效率,Pedro等[83]研究发现,当链霉蛋白酶或DNase Ⅰ与苯扎氯铵(Benzalkonium chloride,BAC) 一起使用时,可增强BAC的杀菌性能,有效降解单核细胞增生李斯特氏菌和大肠杆菌的混合生物被膜。笔者课题组研究发现,酸性电解水能有效清除副溶血性弧菌的生物被膜[84],当其与DNase Ⅰ联合使用时,可增强酸性电解水的清除作用[85]。
3.3 酶制剂与物理杀菌技术联合使用酶制剂与物理杀菌方式结合是一种有效的清除食源性致病菌生物被膜方式。例如,Oulahal-Lagsir等[86]利用淀粉葡萄糖苷酶结合超声波处理,成功清除了大肠杆菌在不锈钢表面的生物被膜。Oulahal等[87]进一步研究了蛋白酶、木瓜蛋白酶、EDTA、超声波的联合处理方式,可有效清除不锈钢表面75%的大肠杆菌生物被膜,而胰蛋白酶、溶菌酶、EDTA、超声波的联合处理方式,可清除不锈钢表面100%的金黄色葡萄球菌生物被膜。
4 总结与展望酶制剂能够靶向作用于QS信号分子、C-di-GMP、胞外多糖、胞外蛋白、eDNA等胞外基质,从根本上清除食源性致病菌的生物被膜,而且因其具有特定的专一性和高效性,使致病菌很难产生相应的耐药性,因此被视为解决食源性致病菌生物被膜危害的重要技术手段之一[14]。本文系统地综述了不同种类的抗生物被膜酶制剂,为食源性致病菌生物被膜的有效控制提供了新的思路和视野。现阶段,酶制剂清除食源性致病菌生物被膜的研究已陆续开展,具有重要的科研价值和广阔的应用前景[12]。但是,抗生物被膜酶制剂在食品工业中的进一步开发与应用,仍存在不少技术难点和上升空间。因此,本文基于抗生物被膜酶制剂的研究现状,对其未来研究方向进行了以下三点展望。
4.1 提高抗生物被膜酶制剂的稳定性酶的专一性和高效性在其清除食源性致病菌生物被膜中发挥着重要作用,但酶的不稳定性可能会影响其在医疗卫生和食品工业中的应用[88]。因此,有效提升酶制剂的稳定性,对其实际的工业化应用至关重要。酶的定向进化技术获得了2018年诺贝尔化学奖[89],该技术可极大地提升酶制剂的稳定性和催化效率。因此,可将该技术应用于抗生物被膜酶制剂的优化,在人工控制的特殊环境下,基于酶的三维结构,通过酶工程技术模拟自然进化机制,定向选择得到具有高稳定性、高清除效力的酶制剂,从而更好地推动抗食源性致病菌生物被膜酶制剂的开发。
4.2 开发基于酶制剂的抗生物被膜生物材料抗生物被膜酶制剂的高效清除能力,使其在开发新型生物材料方面具有良好的应用前景。未来可合理运用固定化酶技术,将酶固定在玻璃、塑料、不锈钢等材料表面,从源头抑制生物被膜的形成,构建一种食源性致病菌生物被膜难以生长的新型生物材料。此外,还可将其与磁珠等纳米材料相结合,提高酶制剂的作用效果,扩大其在食品领域的应用范围。
4.3 促进抗生物被膜酶制剂的工业化抗生物被膜酶制剂是一种绿色环保的技术,但是大多数酶制剂尚处于实验室研究阶段,仅证明了其高效的清除生物被膜能力,具体的工业化应用尚未完全开展。因此,应该利用基因工程技术筛选能够大量表达抗生物被膜酶制剂的高产菌株,提升酶的产量,增强酶的杀菌效果,提高其对食源性致病菌生物被膜的清除效率,推动抗被膜酶制剂在食品工业中的应用。
参考文献
| [1] | World Health Organization. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007-2015. Geneva: WHO, 2015. |
| [2] | O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Ann Rev Microbiolo, 2000, 54: 49-79. DOI:10.1146/annurev.micro.54.1.49 |
| [3] | Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol, 2010, 8(9): 623-633. DOI:10.1038/nrmicro2415 |
| [4] | Chmielewski RAN, Frank JF. Biofilm formation and control in food processing facilities. Comprehens Rev Food Sci Food Safety, 2003, 2(1): 22-32. DOI:10.1111/j.1541-4337.2003.tb00012.x |
| [5] | Mah TFC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol, 2001, 9(1): 34-39. DOI:10.1016/S0966-842X(00)01913-2 |
| [6] | Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev, 2017, 41(3): 276-301. |
| [7] | 李欢. 副溶血性弧菌耐药性微进化机制初步研究[D]. 上海: 上海海洋大学, 2018. Li H. Preliminary research on microevolution mechanisms of antimicrobial resistance of Vibrio parahaemolyticus[D]. Shanghai: Shanghai Ocean University, 2018 (in Chinese). |
| [8] | Chen P, Wang JJ, Ho ng, B, et al. Characterization of mixed-species biofilm formed by Vibrio parahaemolyticus and Listeria monocytogenes. Front Microbiol, 2019, 10: 2543. DOI:10.3389/fmicb.2019.02543 |
| [9] | Srey S, Jahid IK, Ha SD. Biofilm formation in food industries: a food safety concern. Food Control, 2013, 31(2): 572-585. DOI:10.1016/j.foodcont.2012.12.001 |
| [10] | Sim?es M, Sim?es LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT-Food Sci Technol, 2010, 43(4): 573-583. DOI:10.1016/j.lwt.2009.12.008 |
| [11] | Burridge L, Weis JS, Cabello F, et al. Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture, 2010, 306(1/4): 7-23. |
| [12] | Vadivambal R, Jayas DS. Non-uniform temperature distribution during microwave heating of food materials — a review. Food Bioproc Technol, 2010, 3(2): 161-171. DOI:10.1007/s11947-008-0136-0 |
| [13] | Nahar S, Mizan FR, Ha AJW, et al. Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr Rev Food Sci Food Safety, 2018, 17(6): 1484-1502. DOI:10.1111/1541-4337.12382 |
| [14] | Fang KL, Park OJ, Hong SH. Controlling biofilms using synthetic biology approaches. Biotechnol Adv, 2020, 40: 107518. |
| [15] | 赵勇, 李欢, 张昭寰, 等. 食源性致病菌耐药机制研究进展. 生物加工过程, 2018, 16(2): 1-10. Zhao Y, Li H, Zhang ZH, et al. Progress in studying antimicrobial resistance of foodborne pathogenic bacteria. Bioprocess Biosyst Eng, 2018, 16(2): 1-10 (in Chinese). |
| [16] | Zheng Z, Stewart PS. Growth limitation of Staphylococcus epidermidis in biofilms contributes to rifampin tolerance. Biofilms, 2004, 1(1): 31-35. DOI:10.1017/S1479050503001042 |
| [17] | Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol, 2004, 2(2): 95-108. DOI:10.1038/nrmicro821 |
| [18] | Kadariya J, Smith TC, Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed Res Int, 2014, 2014: 827965. |
| [19] | Sharma M, Anand SK. Characterization of constitutive microflora of biofilms in dairy processing lines. Food Microbiol, 2002, 19(6): 627-636. DOI:10.1006/fmic.2002.0472 |
| [20] | 柴旭锋, 齐家伟, 赵莉, 等. 副溶血弧菌在鱼鳞表面形成生物被膜的动态过程及酸性电解水对其清除效果. 上海海洋大学学报, 2019, 28(5): 792-800. Chai XF, Qi JW, Zhao L, et al. Eradication effect of acidic electrolyzed water on Vibrio parahemolyticus biofilm formed on fish scale surface. Journal of Shanghai Ocean University, 2019, 28(5): 792-800 (in Chinese). |
| [21] | 俞文英, 韩乔, 宋雪迎, 等. 冷激条件下预形成副溶血性弧菌生物被膜的发展变化. 微生物学报, 2020, 60(1): 36-48. Yu WY, Han Q, Song XY, et al. Development and changes of biofilm of Vibrio parahaemolyticus preformed under cold excitation. Acta Microbiol Sin, 2020, 60(1): 36-48 (in Chinese). |
| [22] | Song XY, Ma YJ, Fu JJ, et al. Effect of temperature on pathogenic and non-pathogenic Vibrio parahaemolyticus biofilm formation. Food Control, 2017, 73: 485-491. DOI:10.1016/j.foodcont.2016.08.041 |
| [23] | 董旭日, 柴旭锋, 檀玲, 等. 副溶血性弧菌-霍乱弧菌混合生物被膜形成过程研究. 微生物学报, 2018, 58(10): 1808-1816. Dong XR, Chai XF, Tan L, et al. Study on the formation of mixed biofilm of Vibrio parahaemolyticus and Vibrio cholerae. Acta Microbiol Sin, 2018, 58(10): 1808-1816 (in Chinese). |
| [24] | Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Ann Rev Cell Dev Biol, 2005, 21: 319-346. DOI:10.1146/annurev.cellbio.21.012704.131001 |
| [25] | Chbib C. Impact of the structure-activity relationship of AHL analogues on quorum sensing in gram-negative bacteria. Bioorgan Med Chem, 2020, 28(3): 115282. DOI:10.1016/j.bmc.2019.115282 |
| [26] | Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv, 2013, 31(2): 224-245. |
| [27] | Dong YH, Xu JL, Li XZ, et al. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum- sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA, 2000, 97(7): 3526-3531. DOI:10.1073/pnas.97.7.3526 |
| [28] | Chen RD, Zhou ZG, Cao YA, et al. High yield expression of an AHL-lactonase from Bacillus sp. B546 in Pichia pastoris and its application to reduce Aeromonas hydrophila mortality in aquaculture. Microb Cell Fact, 2010, 9: 39. DOI:10.1186/1475-2859-9-39 |
| [29] | Wang WZ, Morohoshi T, Ikenoya M, et al. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl Environ Microbiol, 2010, 76(8): 2524-2530. DOI:10.1128/AEM.02738-09 |
| [30] | Mei GY, Yan XX, Turak A, et al. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl Environ Microbiol, 2010, 76(15): 4933-4942. DOI:10.1128/AEM.00477-10 |
| [31] | Krysciak D, Schmeisser C, Preu? S, et al. Involvement of multiple loci in quorum quenching of autoinducer Ⅰ molecules in the nitrogen-fixing symbiont Rhizobium (Sinorhizobium) sp. Strain NGR234. Appl Environ Microbiol, 2011, 77(15): 5089-5099. DOI:10.1128/AEM.00112-11 |
| [32] | Schipper C, Hornung C, Bijtenhoorn P, et al. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl Environ Microbiol, 2009, 75(1): 224-233. |
| [33] | Lin YH, Xu JL, Hu JY, et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol, 2003, 47(3): 849-860. |
| [34] | Morohoshi T, Nakazawa S, Ebata A, et al. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci Biotechnol Biochem, 2008, 72(7): 1887-1893. |
| [35] | Chen CN, Chen CJ, Liao CT, et al. A probable aculeacin A acylase from the Ralstonia solanacearum GMI1000 is N-acyl-homoserine lactone acylase with quorum-quenching activity. BMC Microbiol, 2009, 9: 89. |
| [36] | Wahjudi M, Papaioannou E, Hendrawati O, et al. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology, 2011, 157(7): 2042-2055. |
| [37] | Sio CF, Otten LG, Cool RH, et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun, 2006, 74(3): 1673-1682. |
| [38] | Bijtenhoorn P, Mayerhofer H, Müller-Dieckmann J, et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE, 2011, 6(10): e26278. |
| [39] | Chan KG, Atkinson S, Mathee K, et al. Characterization of N-acylhomoserine lactone- degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol, 2011, 11: 51. |
| [40] | Chowdhary PK, Keshavan N, Nguyen HQ, et al. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry, 2007, 46(50): 14429-14437. |
| [41] | Uroz S, Chhabra SR, Cámara M, et al. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology, 2005, 151(10): 3313-3322. |
| [42] | Ryjenkov DA, Tarutina M, Moskvin OV, et al. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol, 2005, 187(5): 1792-1798. |
| [43] | Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA, 2005, 102(40): 14422-14427. |
| [44] | Wassmann P, Chan C, Paul R, et al. Structure of BeF3– -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure, 2007, 15(8): 915-927. |
| [45] | De N, Pirruccello M, Krasteva PV, et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol, 2008, 6(3): e67. |
| [46] | Chen LH, Koseoglu VK, Güvener ZT, et al. Cyclic di-GMP-dependent signaling pathways in the pathogenic firmicute Listeria monocytogenes. PLoS Pathog, 2014, 10(8): e1004301. |
| [47] | Minasov G, Padavattan S, Shuvalova L, et al. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. Journal of Biological Chemistry, 2009, 284(19): 13174-13184. |
| [48] | Barends TRM, Hartmann E, Griese JJ, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature, 2009, 459(7249): 1015-1018. |
| [49] | Lovering AL, Capeness MJ, Lambert C, et al. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio, 2011, 2(5): e00163-11. |
| [50] | Rybtke M, Hultqvist LD, Givskov M, et al. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol, 2015, 427(23): 3628-3645. |
| [51] | Colvin KM, Gordon VD, Murakami K, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog, 2011, 7(1): e1001264. |
| [52] | Li W, Wang JJ, Qian H, et al. Insights into the role of extracellular DNA and extracellular proteins in biofilm formation of Vibrio parahaemolyticus. Front Microbiol, 2020, 11: 813. |
| [53] | Guo LX, Wang JJ, Gou Y, et al. Comparative proteomics reveals stress responses of Vibrio parahaemolyticus biofilm on different surfaces: internal adaptation and external adjustment. Sci Total Environ, 2020, 731: 138386. |
| [54] | Wang YJ, Moradali MF, Goudarztalejerdi A, et al. Biological function of a polysaccharide degrading enzyme in the periplasm. Sci Rep, 2016, 6: 31249. |
| [55] | Baker P, Hill PJ, Snarr BD, et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv, 2016, 2(5): e1501632. |
| [56] | Shen WR, Zhao XR, Wang XL, et al. Improving the power generation performances of gram-positive electricigens by regulating the peptidoglycan layer with lysozyme. Environ Res, 2020, 185: 109463. |
| [57] | Fleming D, Chahin L, Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother, 2017, 61(2): e01998-16. |
| [58] | Izano EA, Wang H, Ragunath C, et al. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dental Res, 2007, 86(7): 618-622. |
| [59] | Leroy C, Delbarre C, Ghillebaert F, et al. Influence of subtilisin on the adhesion of a marine bacterium which produces mainly proteins as extracellular polymers. J Appl Microbiol, 2008, 105(3): 791-799. |
| [60] | Ceotto-Vigoder H, Marques SLS, Santos INS, et al. Nisin and lysostaphin activity against preformed biofilm of Staphylococcus aureus involved in bovine mastitis. J Appl Microbiol, 2016, 121(1): 101-114. |
| [61] | Gutierrez D, Briers Y, Rodriguez-Rubio L, et al. Role of the pre-neck appendage protein (Dpo7) from phage Vb_SepiS-phiIPLA7 as an anti-biofilm agent in staphylococcal species. Front Microbiol, 2015, 6: 1315. |
| [62] | Thomas VC, Thurlow LR, Boyle D, et al. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol, 2008, 190(16): 5690-5698. |
| [63] | Franklin MJ, Nivens DE, Weadge JT, et al. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol, 2011, 2: 167. |
| [64] | Strathmann M, Wingender J, Flemming HC. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J Microbiol Methods, 2002, 50(3): 237-248. |
| [65] | Lamppa JW, Ackerman ME, Lai JI, et al. Genetically engineered alginate lyase-PEG conjugates exhibit enhanced catalytic function and reduced immunoreactivity. PLoS ONE, 2011, 6(2): e17042. |
| [66] | Mai GT, Seow WK, Pier GB, et al. Suppression of lymphocyte and neutrophil functions by Pseudomonas aeruginosa mucoid exopolysaccharide (alginate): reversal by physicochemical, alginase, and specific monoclonal antibody treatments. Infection and immunity, 1993, 61(2): 559-564. |
| [67] | Asker D, Awad TS, Baker P, et al. Non-eluting, surface-bound enzymes disrupt surface attachment of bacteria by continuous biofilm polysaccharide degradation. Biomaterials, 2018, 167: 168-176. |
| [68] | Earl C, Arnaouteli S, Bamford NC, et al. The majority of the matrix protein TapA is dispensable for Bacillus subtilis colony biofilm architecture. Mol Microbiol, 2020, 114(6): 920-933. |
| [69] | Raulinaitis V, Tossavainen H, Aitio O, et al. Identification and structural characterization of LytU, a unique peptidoglycan endopeptidase from the lysostaphin family. Sci Rep, 2017, 7: 6020. |
| [70] | Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol, 2015, 41(3): 341-352. |
| [71] | Zetzmann M, Okshevsky M, Endres J, et al. DNase-sensitive and-resistant modes of biofilm formation by Listeria monocytogenes. Front Microbiol, 2015, 6: 1428. |
| [72] | Chiang WC, Nilsson M, Jensen P?, et al. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother, 2013, 57(5): 2352-2361. |
| [73] | Schwartz K, Ganesan M, Payne DE, et al. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol Microbiol, 2013, 99(1): 123-134. |
| [74] | Tetz GV, Artemenko NK, Tetz VV. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother, 2009, 53(3): 1204-1209. |
| [75] | Karygianni L, Attin T, Thurnheer T. Combined DNase and proteinase treatment interferes with composition and structural integrity of multispecies oral biofilms. J Clin Med, 2020, 9(4): 983. |
| [76] | Thallinger B, Prasetyo EN, Nyanhongo GS, et al. Antimicrobial enzymes: an emerging strategy to fight microbes and microbial biofilms. Biotechnol J, 2013, 8(1): 97-109. |
| [77] | Thallinger B, Brandauer M, Burger P, et al. Cellobiose dehydrogenase functionalized urinary catheter as novel antibiofilm system. J Biomed Mater Res Part B: Appl Biomater, 2016, 104(7): 1448-1456. |
| [78] | Zanaroli G, Negroni A, Calisti C, et al. Selection of commercial hydrolytic enzymes with potential antifouling activity in marine environments. Enzyme Microb Technol, 2011, 49(6/7): 574-579. |
| [79] | Kiran GS, Lipton AN, Kennedy J, et al. A halotolerant thermostable lipase from the marine bacterium Oceanobacillus sp. PUMB02 with an ability to disrupt bacterial biofilms. Bioengineered, 2014, 5(5): 305-318. |
| [80] | Nguyen UT, Burrows LL. DNase Ⅰ and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int J Food Microbiol, 2014, 187: 26-32. |
| [81] | Johansen C, Falholt P, Gram L. Enzymatic removal and disinfection of bacterial biofilms. Appl Environ Microbiol, 1997, 63(9): 3724-3728. |
| [82] | Walker SL, Fourgialakis M, Cerezo B, et al. Removal of Microbial biofilms from dispense equipment: the effect of enzymatic pre-digestion and detergent treatment. J Instit Brew, 2007, 113(1): 61-66. |
| [83] | Rodríguez-López P, Carballo-Justo A, Draper LA, et al. Removal of Listeria monocytogenes dual-species biofilms using combined enzyme-benzalkonium chloride treatments. Biofouling, 2017, 33(1): 45-58. |
| [84] | Han Q, Song XY, Zhang ZH, et al. Removal of foodborne pathogen biofilms by acidic electrolyzed water. Front Microbiol, 2017, 8: 988. |
| [85] | Li YF, Tan L, Guo LX, et al. Acidic electrolyzed water more effectively breaks down mature Vibrio parahaemolyticus biofilm than DNase I. Food Control, 2020, 117: 107312. |
| [86] | Oulahal-Lagsir O, Martial-Gros A, Bonneau M, et al. "Escherichia coli-milk" biofilm removal from stainless steel surfaces: synergism between ultrasonic waves and enzymes. Biofouling, 2003, 19(3): 159-168. |
| [87] | Oulahal N, Martial-Gros A, Bonneau M, et al. Removal of meat biofilms from surfaces by ultrasounds combined with enzymes and/or a chelating agent. Innovat Food Sci Emerg Technol, 2006, 8(2): 192-196. |
| [88] | Wang QF, Nie P, Hou YH, et al. . Purification, biochemical characterization and DNA protection against oxidative damage of a novel recombinant superoxide dismutase from psychrophilic bacterium Halomonas sp. ANT108. Prot Express Purificat, 2020, 173: 105661. |
| [89] | Chowdhury R, Maranas CD. From directed evolution to computational enzyme engineering — a review. AIChE J, 2020, 66(3): e16847. |
