
 , 王松涛4, 沈才洪4, 史劲松3, 许正宏1,2,4
, 王松涛4, 沈才洪4, 史劲松3, 许正宏1,2,4

1. 江南大学 生物工程学院 工业生物技术教育部重点实验室,江苏 无锡 214122;
2. 江南大学 粮食发酵工艺与技术国家工程实验室,江苏 无锡 214122;
3. 江南大学 江苏省生物活性制品加工工程技术研究中心,江苏 无锡 214122;
4. 国家固态酿造工程技术研究中心,四川 泸州 646000;
5. 苏州科技大学 化学生物与材料工程学院,江苏 苏州 215009
收稿日期:2019-10-11;接收日期:2019-12-03;网络出版时间:2019-12-09
基金项目:国家自然科学基金(Nos. 31771967, 31530055, 31901658),国家轻工技术与工程一流学科自主课题(No. LITE2018-11)资助
通讯作者:Qi Li. Tel: +86-510-85918176, E-mail: liqi@jiangnan.edu.cn;
Zhenghong Xu. Tel: +86-510-85918206, E-mail: zhenghxu@jiangnan.edu.cn.
摘要:梭菌对泸型酒风味品质的发酵形成具有重要作用,但目前对酒醅和窖泥中梭菌群落的物种组成、发酵演替规律以及代谢功能的差异尚缺少深入认识。采用分子微生态学技术,在种水平上对比了同一窖池的酒醅和窖泥中梭菌纲细菌群落的结构差异和演替规律,并通过纯培养方法分离和评价了梭菌菌株的主要挥发性脂肪酸组成差异。结果表明,窖泥发酵过程中梭菌纲细菌和总细菌的基因拷贝数比值相对稳定(71.5%–91.2%),而酒醅中梭菌的变化则较大(0.9%–36.5%)。酒醅中优势梭菌纲细菌主要是梭菌属(Clostridium,19.9%)、沉积物菌属(Sedimentibacter,8.8%)和氢孢菌属(Hydrogenispora,7.2%),而窖泥中主要为氢孢菌属(57.2%)、沉积物菌属(5.4%)和产己酸菌属(Caproiciproducens,4.9%)。窖泥及酒醅发酵过程中梭菌群落结构差异显著(P=0.001)。分离获得的20株梭菌菌株具有不同的产挥发性脂肪酸能力。结果表明,窖池中梭菌纲细菌群落存在时空异质性,其结构和功能差异对泸型酒风味形成具有影响。
关键词:浓香型白酒酒醅窖泥微生物群落梭菌纲细菌
Differences of the structure, succession and function of Clostridial communities between jiupei and pit mud during Luzhou-flavour baijiu fermentation
Wei Qian1,5, Zhenming Lu2,4, Lijuan Chai2, Xiaojuan Zhang2,4, Pengxiang Xu1, Qi Li1

 , Songtao Wang4, Caihong Shen4, Jinsong Shi3, Zhenghong Xu1,2,4
, Songtao Wang4, Caihong Shen4, Jinsong Shi3, Zhenghong Xu1,2,4

1. Key Laboratory of Industrial Biotechnology of Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu, China;
2. National Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi 214122, Jiangsu, China;
3. Jiangsu Engineering Research Center for Bioactive Products Processing Technology, Jiangnan University, Wuxi 214122, Jiangsu, China;
4. National Engineering Research Center of Solid-State Brewing, Luzhou 646000, Sichuan, China;
5. School of Chemistry, Biology and Materials Engineering, Suzhou University of Science and Technology, Suzhou 215009, Jiangsu, China
Received: October 11, 2019; Accepted: December 3, 2019; Published: December 9, 2019
Supported by: National Natural Science Foundation of China (Nos. 31771967, 31530055, 31901658), National First-class Discipline Program of Light Industry Technology and Engineering (No. LITE2018-11)
Abstract: Clostridia inhabiting in jiupei and pit mud plays key roles in the formation of flavour during the fermentation process of Luzhou-flavour baijiu. However, the differences of Clostridial communities between jiupei and pit mud remains unclear. Here, the species assembly, succession, and metabolic capacity of Clostridial communities between jiupei and pit mud were analysed by high-throughput sequencing and pure culture approaches. The ratio of Clostridial biomass to bacterial biomass in the pit mud was relatively stable (71.5%–91.2%) throughout the fermentation process. However, it varied widely in jiupei (0.9%–36.5%). The dominant Clostridial bacteria in jiupei were Clostridium (19.9%), Sedimentibacter (8.8%), and Hydrogenispora (7.2%), while Hydrogenispora (57.2%), Sedimentibacter (5.4%), and Caproiciproducens (4.9%) dominated in the Clostridial communities in pit mud. The structures of Clostridial community in pit mud and jiupei were significantly different (P=0.001) throughout fermentation. Isolated Clostridial strains showed different metabolic capacities of volatile fatty acids in pure culture. Spatial and temporal heterogeneity of Clostridial communities existed in the baijiu fermentation pit, which was closely related to the main flavour components of Luzhou-flavour baijiu.
Keywords: strong-flavour baijiujiupeipit mudmicrobial communityClostridia
泸型酒是浓香型白酒的典型代表,具有独特的风格和口味,深受广大消费者喜爱。泥窖是泸型酒酿造过程中使用的一种独特生物反应器,其特有的营养环境条件为种类繁多的微生物提供了栖息繁衍场所[1]。梭菌纲细菌(Clostridia)是窖池微生态系统的重要组成,也是酿酒过程产生己酸、丁酸等风味物质的主要功能微生物[2],窖泥中存在的多种梭菌还能够与甲烷菌共栖,产生独特的风味代谢产物[3-4]。已有研究表明,酒醅和窖泥里面均存在种类多样的梭菌,但目前对酒醅和窖泥中梭菌的种类、发酵演替规律以及酿造代谢功能的差异尚缺少系统比较。
本研究以连续使用的泸州老窖优质百年窖池内酒醅和窖泥中梭菌群落为研究对象,通过分子生态学技术分析对比了酿造过程酒醅和窖泥中梭菌纲细菌群落的物种多样性和发酵演替规律,并对纯培养分离的梭菌菌株发酵产生短链脂肪酸特性进行了对比分析,为理解梭菌在泸型白酒酿造过程中的作用提供理论基础。
1 材料与方法1.1 样品采集样品采自四川泸州老窖罗汉酿酒基地100年窖龄窖池(长×宽×高=4.3 m×2.3 m×2.3 m)。自入窖开始第0、1、3、5、7、9、11、16、21、26、36、46天(发酵结束),使用自制取样工具于窖池中层采集酒醅样品,于窖池底部采集窖泥样品,每份约50 g,每个时间点取3个平行样[5]。取样样品迅速装入无菌塑封袋,?80 ℃保存。
1.2 主要仪器和材料Whitley DG250厌氧培养箱,英国Don Whitley Scientific公司;ND 2000核酸检测仪,美国Thermo Fisher Scientific公司;荧光定量PCR仪Bio-Rad CFX Real-Time System,美国Bio-Rad公司;Illumina HiSeq 2000测序平台,深圳华大基因;PowerSoil? DNA提取试剂盒,美国MOBIO公司;乙酸、丁酸、戊酸、己酸标准品,上海阿拉丁试剂。
梭菌强化培养基(RCM,g/L):葡萄糖5,胰蛋白胨10,酵母粉3,牛肉膏10,氯化钠3,无水乙酸钠3,L-半胱氨酸盐酸盐0.5,可溶性淀粉1,pH 6.5,121 ℃下高压灭菌20 min。ES培养基(g/L):乙酸钠5,酵母粉1,硫酸镁0.2,磷酸氢二钾0.4,硫酸铵0.5,pH 6.0,121 ℃高压灭菌20 min后加入无水乙醇(终浓度20 mL/L)。
1.3 宏基因组DNA提取称取5.0 g样品至无菌陶瓷研钵中,加入液氮研磨。每份样品称取0.2 g粉末,采用试剂盒提取DNA,检测DNA浓度和纯度(A260/A280值在1.8–2.0之间)。
1.4 总细菌和梭菌纲细菌定量分析采用荧光定量PCR (qPCR),对酒醅和窖泥中总细菌和梭菌纲细菌进行定量分析[5-6]。使用细菌通用引物338F/518R[7]和梭菌纲细菌特异性引物SJ-F/SJ-R[8],以Cq值为横坐标,Lg (copies/g)活菌数为纵坐标,绘制大肠杆菌Escherichia coli生物量标准曲线Y=?0.341X+12.106 (R2=0.995 6) (图 1A)和克氏梭菌Clostridium kluyveri生物量标准曲线Y=?0.285X+11.664 (R2=0.996 3) (图 1B)。
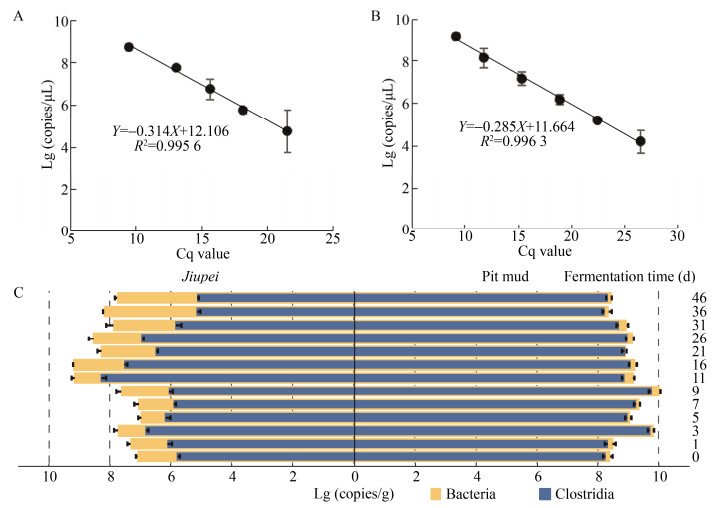 |
| 图 1 发酵过程酒醅和窖泥中总细菌及梭菌纲细菌的实时荧光定量结果 Fig. 1 The Real-time quantitative PCR results of bacteria and Clostridia in jiupei and pit mud during fermentation. |
| 图选项 |
1.5 16S rRNA扩增子测序分析采用细菌通用引物338F/806R扩增16S rRNA基因的V3/V4区[9],构建测序文库后进行高通量测序。采用QIIME (V1.9.1)对测序原始数据进行质控与分析,以97%相似度为标准对测序序列进行操作分类单元(OTU)划分[10-11],并使用SILVA和RDP数据库对OTU进行分类学注释。分析Shannon、Chao1、Ace等微生物群落α-多样性指数[12]。采用R (version 3.4.4)对各样本进行主坐标分析(PCoA)及聚类分析。采用LEfSe (LDA Effect Size)分析对比不同样本组间差异菌群[13]。测序数据提交至美国国立生物技术信息中心SRA数据库(http://www.ncbi.nlm.nih.gov/traces/sra),登录号为PRJNA558446。
1.6 梭菌的分离鉴定及代谢产物检测取5.0 g酒醅或窖泥分别加入100 mL无菌生理盐水,2 000 r/min振荡5 min,80 ℃热处理10 min后,以5%接种量接种至厌氧预处理的RCM和ES液体培养基中,厌氧(N2:H2:CO2= 8:1:1)培养14 d。培养液稀释涂布于RCM和ES固体培养基,挑取单菌落进行液体培养7 d,离心收集菌体后采用试剂盒提取DNA,利用细菌通用引物27F/1492R[14]扩增16S rRNA基因序列后进行测序[15]。梭菌菌株接种至RCM液体培养基,厌氧静置培养7 d后,检测发酵液中短链脂肪酸(乙酸、丁酸、戊酸和己酸等)含量[16]。
2 结果与分析2.1 总细菌及梭菌纲细菌的定量分析整个发酵过程中,酒醅中梭菌和总细菌的基因拷贝数比值变化较大(0.9%–36.5%),而窖泥中梭菌和总细菌的基因拷贝数比值则相对比较稳定(71.5%–91.2%)。酒醅中梭菌的基因拷贝数在第11天升至最高(1.5×109 copies/g),然后迅速降低,发酵结束时(46 d)为1.3×107 copies/g;窖泥中总细菌、梭菌纲细菌均呈现先增加后减少趋势,第9天达最高(1.0×1010 copies/g和7.8×109 copies/g),随后逐渐下降并保持相对稳定(图 1C)。
2.2 梭菌群落的α-多样性窖泥和酒醅样本高通量测序共获得963个梭菌纲OTU,平均覆盖度分别为99.1%和89.2%。酒醅梭菌群落Chao1丰富度指数在发酵前3天缓慢上升至346.7,随后不断下降至发酵结束的43.5;窖泥梭菌群落Chao1指数(493.0–630.9)明显高于酒醅。随着发酵进行,酒醅中梭菌群落的Shannon多样性指数从6.3下降为4.0,而窖泥梭菌群落多样性指数在3.6–5.8之间动态变化。
2.3 窖池中梭菌群落的发酵演替规律酒醅中梭菌纲细菌主要由梭菌科(Clostridiaceae 1,占梭菌纲平均相对丰度23.7%)、毛螺菌科(Lachnospiraceae,17.4%)、Clostridiales Family Ⅺ (16.5%)、瘤胃菌科(Ruminococcaceae,15.7%)和太阳杆菌科(Heliobacteriaceae,7.9%)组成,而窖泥中优势种群是太阳杆菌科(57.2%)、瘤胃菌科(10.4%)和Clostridiales Family Ⅺ (6.6%) (图 2)。
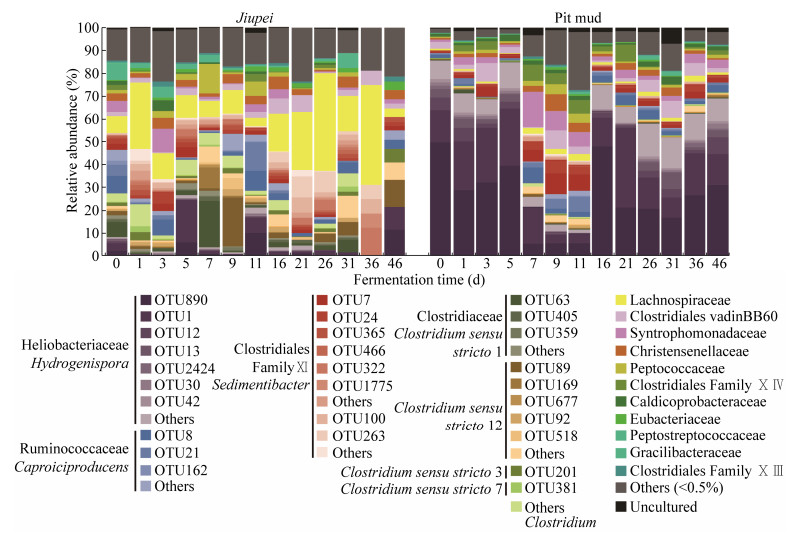 |
| 图 2 酒醅和窖泥中梭菌群落的演替规律 Fig. 2 Succession of Clostridial community in jiupei and pit mud throughout fermentation. |
| 图选项 |
发酵开始时,氢孢菌属(OTU890、OTU1)是窖泥梭菌群落的优势物种(> 80%),但在酒醅中相对丰度低于10%;发酵7–11 d,窖泥中产己酸菌属 (OTU8、OTU21)和沉积物菌属 (OTU7、OTU24)等物种的相对丰度较发酵初期(0–5 d)有所增加,均值分别为9.84%和12.97%,而酒醅中梭菌属的相对丰度变化相对较小;发酵16–46 d,窖泥梭菌群落结构逐渐恢复至发酵初始结构,酒醅中毛螺菌科逐渐增长为优势类群,36 d时相对丰度达43.75% (图 2)。
2.4 窖泥与酒醅中梭菌群落结构的差异对种水平的梭菌纲细菌群落组成进行PCoA分析,结果表明酒醅和窖泥在第一轴(占总方差的40.7%)上可以明显被分为两组,且两者群落结构差异显著(R=0.65,P=0.001) (图 3A)。
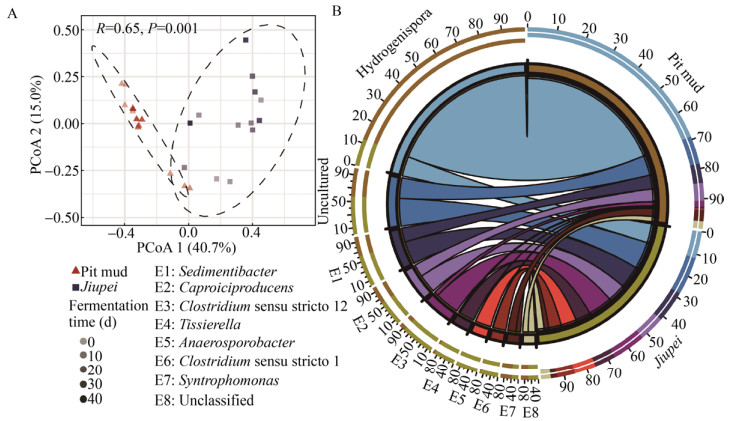 |
| 图 3 窖泥与酒醅梭菌纲细菌群落结构比较 Fig. 3 Structure of Clostridial communities in jiupei and pit mud. (A) The PCoA scores plot. (B) The Circos plot of relative abundance of Clostridia (genus level). |
| 图选项 |
窖池中氢孢菌属(Hydrogenispora)和互营单胞菌属(Syntrophomonas)主要分布在窖泥中,其相对丰度分别是酒醅中的7.6倍和1.7倍;而窖池中厌氧孢杆菌属(Anaerosporobacter)、狭义梭菌属1 (Clostridium sensu stricto 1)和组织菌属(Tissierella)主要分布在酒醅样品中,其丰度分别是窖泥中的74.8、18.6和10.2倍(图 3B)。
2.5 梭菌纲细菌的筛选及代谢产物分析采用平板分离和16S rRNA基因测序的方法,从酒醅和窖泥样品中共分离和鉴定了54株梭菌,分为4个科7个属20个种。每种梭菌选取1株代表菌株,在RCM液体培养基中厌氧静置培养7 d后,检测发酵液中的主要短链脂肪酸(乙酸、丁酸、戊酸、己酸)的组成,结果见表 1。
表 1 梭菌菌株发酵液中主要脂肪酸组成Table 1 Production of main fatty acids in the fermentation broth of Clostridial species
| ID | Taxonomy | Medium | GenBankID value | Fatty acid production (g/L) | |||
| Acetic acid | Butyric acid | Valeric acid | Hexanoic acid | ||||
| Mui03 | Muricomes intestini | RCM | 99.5% | 2.2±0.0 | 0.4±0.0 | 2.0±0.1 | 0.6±0.0 |
| Car01 | Clostridium carboxidivorans | RCM | 99.4% | 1.8±0.0 | 0.9±0.1 | 4.1±0.5 | 0.9±0.0 |
| Swc01 | Clostridium swellfunianum | ES | 96.9% | 2.3±0.1 | 1.1±0.0 | 1.9±0.2 | 1.3±0.1 |
| Sca01 | Clostridium scatologenes | RCM | 99.9% | 2.2±0.1 | 1.2±0.2 | ND | 1.3±0.1 |
| Tep01 | Terrisporobacter petrolearius | RCM | 99.9% | 2.0±0.1 | 0.2±0.1 | 1.7±0.0 | 0.1±0.0 |
| Clba01 | Clostridium pasteurianum | RCM | 99.9% | 1.9±0.1 | 0.9±0.1 | 1.7±0.0 | 0.8±0.0 |
| Clau01 | Clostridium autoethanogenum | RCM | 99.2% | 1.8±0.1 | ND | 2.6±0.2 | 0.9±0.0 |
| Claci01 | Clostridium aciditolerans | FWM | 99.6% | 1.3±0.0 | 0.9±0.7 | 0.4±0.1 | ND |
| Cko03 | Clostridium kogasensis | RCM | 99.3% | 1.5±0.3 | 1.5±0.1 | 2.5±0.1 | 0.7±0.1 |
| Cam01 | Clostridium amylolyticum | RCM | 100.0% | 2.3±0.0 | ND | 2.0±0.0 | 0.9±0.0 |
| Cls01 | Clostridium senegalense | RCM | 100.0% | 3.0±0.0 | 0.8±0.2 | 2.2±0.1 | 0.9±0.0 |
| Clu05 | Clostridium luticellarii | ES | 99.6% | 1.0±0.1 | 1.1±0.5 | 1.6±0.0 | 0.7±0.0 |
| Clb01 | Clostridium beijerinckii | RCM | 100.0% | 0.5±0.0 | 2.0±0.1 | 2.1±0.3 | 0.9±0.1 |
| Cn01 | Clostridium novyi | RCM | 99.6% | 2.9±0.3 | 0.6±0.0 | 2.0±0.2 | 0.8±0.1 |
| Clk07 | Clostridium kluyveri | ES | 98.9% | 0.7±0.1 | 1.6±0.2 | 2.0±0.2 | 0.6±0.1 |
| Clbo01 | Clostridium botulinum | RCM | 100.0% | 1.0±0.0 | 0.5±0.1 | 1.9±0.2 | 2.6±0.2 |
| Lab01 | Lachnospiraceae bacterium | ES | 99.6% | 2.4±0.1 | 0.3±0.1 | ND | ND |
| JN500902 | Syntrophococcus sp. | RCM | 93.4% | 2.7±0.0 | 0.2±0.1 | 2.8±0.0 | 1.2±0.0 |
| Alb01 | Alkalibaculum bacchi | RCM | 99.3% | 1.2±0.3 | ND | 1.8±0.0 | 0.4±0.1 |
| Cag01 | Caproiciproducens galactitolivorans | ES | 98.1% | 1.3±0.0 | 0.4±0.1 | 1.8±0.0 | 0.5±0.0 |
| ND: not detected. | |||||||
表选项
所有20株所筛菌株均产乙酸,其中C. senegalense Cls01培养液中乙酸含量可达(3.0±0.0) g/L。20株菌中有17株菌株培养液中检出丁酸,其中拜氏梭菌C. beijerinckii Clb01发酵液丁酸含量最高,为(2.0±0.1) g/L。有18株菌株的培养液中检出戊酸,其中厌氧食气梭菌C. carboxidivorans Car01培养液中戊酸含量最高,达(4.1±0.5) g/L。18株菌株的发酵液中检出己酸,其中肉毒梭菌C. botulinum Clbo01培养液中己酸含量最高,为(2.6±0.2) g/L,而半乳糖已酸菌Caproiciproducens galactitolivorans Cag01培养液中己酸含量为(0.5±0.1) g/L。
3 讨论与结论本研究从种水平对比了同一窖池的酒醅和窖泥中梭菌纲细菌群落的结构差异,结果表明酒醅和窖泥中梭菌群落的结构存在显著差异。酒醅中主要梭菌为梭菌属,且种类较多,而窖泥中主要为氢孢菌属 (OTU890、OTU1)和产己酸菌属 (OTU8、OTU21)。已有研究表明,Hydrogenispora ethanolica LX-BT可利用葡萄糖产乙酸、乙醇和氢气[17];Caproiciproducens galactitolivorans BS-1T在乙酸存在的情况下可加快利用碳源转化为乙醇、乙酸、丁酸和己酸,同时产生H2和CO2[18],与其他产挥发性脂肪酸的菌株共培养时,己酸含量会增加[19];Ruminiclostridium papyrosolvens DSM 2782T能够降解纤维素产生乙醇、乳酸、乙酸、H2和CO2[20-21];Sedimentibacter acidaminivorans MO-SEDIT可利用赖氨酸和甘氨酸为氮源进行Stickland反应,产生乙酸和丁酸[22]。C. carboxidivorans DSM 15243T在H2、CO和CO2存在的情况下,不需利用其他碳源即可发酵产生乙醇、丁醇和己醇[23]。酒醅和窖泥中梭菌种类的差异可能与所处的营养环境有关,这些微生物在发酵过程中演替的机理尚需要进一步解析。
从窖池中分离了20种梭菌菌株,并通过纯培养发现同菌株发酵液中短链脂肪酸组成具有差异性,一方面反映了酒窖中存在的梭菌物种多样性,另一方面也验证了不同梭菌物种的代谢功能差异性。需要说明的是,梭菌菌株在不同培养条件下的代谢特性可能具有明显差异,在窖池发酵过程中梭菌还与其他微生物和环境间存在着多种相互作用关系,其发酵机理还需进一步阐明[24]。与测序结果相比,窖池中另外两个主要类群的梭菌(氢孢菌属和沉积物菌属)尚未分离出纯培养菌株,后续可以利用测序信息指导培养基的选择和设计[25],提高窖池梭菌的可培养性。
本研究利用分子生态学技术分析了泸型酒泥窖发酵过程中酒醅和窖泥的梭菌纲群落结构组成差异和演替规律,采用纯培养方法分离并评价了梭菌纲菌株主要挥发性脂肪酸的组成特征,为解析梭菌群落的酿造功能提供了有益信息。
参考文献
| [1] | Zhang SY, Xu DF, Luzhou Laojiao Group CO., LTD. The Brewing Technique Pandect of Lu-Type Liquor. Beijing: China Light Industry Press, 2011: 89-92 (in Chinese). 张宿义, 许德富, 泸州老窖集团有限责任公司. 泸型酒技艺大全. 北京: 中国轻工业出版社, 2011: 89-92. |
| [2] | Liu MK, Tang YM, Guo XJ, et al. Deep sequencing reveals high bacterial diversity and phylogenetic novelty in pit mud from Luzhou Laojiao cellars for Chinese strong-flavor Baijiu. Food Res Int, 2017, 102: 68-76. DOI:10.1016/j.foodres.2017.09.075 |
| [3] | Zou W, Ye GB, Zhang KZ. Diversity, function, and application of Clostridium in Chinese Strong Flavor Baijiu ecosystem: a review. J Food Sci, 2018, 83(5): 1193-1199. DOI:10.1111/1750-3841.14134 |
| [4] | Wu YY, Lu SH, Liu GY, et al. A study of enhancing the quality of Luzhou type qujiu by using simultaneous fermentation of caproic acid bacteria and methane bacteria. Food Ferment Ind, 1990(6): 1-6 (in Chinese). 吴衍庸, 卢世珩, 刘光烨, 等. 利用窖泥甲烷菌与己酸菌二元发酵技术提高泸型曲酒质量的研究. 食品与发酵工业, 1990(6): 1-6. |
| [5] | Chai LJ, Xu PX, Qian W, et al. Profiling the Clostridia with butyrate-producing potential in the mud of Chinese liquor fermentation cellar. Int J Food Microbiol, 2019, 297: 41-50. DOI:10.1016/j.ijfoodmicro.2019.02.023 |
| [6] | Xiao C, Lu ZM, Zhang XJ, et al. Bacterial community succession in fermented grains of Luzhou-flavor baijiu. Acta Microbiol Sin, 2019, 59(1): 195-204 (in Chinese). 肖辰, 陆震鸣, 张晓娟, 等. 泸型酒酒醅细菌群落的发酵演替规律. 微生物学报, 2019, 59(1): 195-204. |
| [7] | Dennis KL, Wang YW, Blatner NR, et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res, 2013, 73(19): 5905-5913. DOI:10.1158/0008-5472.CAN-13-1511 |
| [8] | Hu XL, Wang HY, Wu Q, et al. Development, validation and application of specific primers for analyzing the clostridial diversity in dark fermentation pit mud by PCR-DGGE. Bioresour Technol, 2014, 163: 40-47. DOI:10.1016/j.biortech.2014.04.008 |
| [9] | Ma J, Lu ZM, Zhang XJ, et al. Role of Clostridia community in fermented grains during Luzhou-flavor liquor production. Microbiol China, 2016, 43(10): 2322 (in Chinese). 马箭, 陆震鸣, 张晓娟, 等. 泸型酒酒醅中梭菌群落的发酵演替规律和功能预测. 微生物学通报, 2016, 43(10): 2322-2329. |
| [10] | Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [11] | Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 2010, 26(19): 2460-2461. DOI:10.1093/bioinformatics/btq461 |
| [12] | Huang YH, Yi ZL, Jin YL, et al. New microbial resource: microbial diversity, function and dynamics in Chinese liquor starter. Sci Rep, 2017, 7(1): 14577. DOI:10.1038/s41598-017-14968-8 |
| [13] | Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol, 2011, 12(6): R60. DOI:10.1186/gb-2011-12-6-r60 |
| [14] | Zhang JR, Fang F, Chen J, et al. The arginine deiminase pathway of koji bacteria is involved in ethyl carbamate precursor production in soy sauce. FEMS Microbiol Lett, 2014, 358(1): 91-97. DOI:10.1111/1574-6968.12542 |
| [15] | Kim OS, Cho YJ, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol, 2012, 62(3): 716-721. |
| [16] | Chai LJ, Lu ZM, Zhang XJ, et al. Zooming in on butyrate-producing Clostridial consortia in the fermented grains of Baijiu via gene sequence-guided microbial isolation. Front Microbiol, 2019, 10: 1397. DOI:10.3389/fmicb.2019.01397 |
| [17] | Liu Y, Qiao JT, Yuan XZ, et al. Hydrogenispora ethanolica gen. nov., sp. nov., an anaerobic carbohydrate-fermenting bacterium from anaerobic sludge. Int J Syst Evol Microbiol, 2014, 64(5): 1756-1762. |
| [18] | Kim BC, Jeon BS, Kim S, et al. Caproiciproducens galactitolivorans gen. nov., sp. nov., a bacterium capable of producing caproic acid from galactitol, isolated from a wastewater treatment plant. Int J Syst Evol Microbiol, 2015, 65(12): 4902-4098. |
| [19] | Jeon BS, Kim BC, Um YS, et al. Production of hexanoic acid from D-galactitol by a newly isolated Clostridium sp. BS-1. Appl Microbiol Biotechnol, 2010, 88(5): 1161-1167. DOI:10.1007/s00253-010-2827-5 |
| [20] | Madden RH, Bryder MJ, Poole NJ. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium papyrosolvens sp. nov. Int J Syst Bacteriol, 1982, 32(1): 87-91. DOI:10.1099/00207713-32-1-87 |
| [21] | Zhang X, Tu B, Dai LR, et al. Petroclostridium xylanilyticum gen. nov., sp. nov., a xylan-degrading bacterium isolated from an oilfield, and reclassification of clostridial cluster Ⅲ members into four novel genera in a new Hungateiclostridiaceae fam. nov. Int J Syst Evol Microbiol, 2018, 68(10): 3197-3211. DOI:10.1099/ijsem.0.002966 |
| [22] | Imachi H, Sakai S, Kubota T, et al. Sedimentibacter acidaminivorans sp. nov., an anaerobic, amino-acid-utilizing bacterium isolated from marine subsurface sediment. Int J Syst Evol Microbiol, 2016, 66(3): 1293-1300. DOI:10.1099/ijsem.0.000878 |
| [23] | Phillips JR, Atiyeh HK, Tanner RS, et al. Butanol and hexanol production in Clostridium carboxidivorans syngas fermentation: medium development and culture techniques. Bioresour Technol, 2015, 190: 114-121. DOI:10.1016/j.biortech.2015.04.043 |
| [24] | Ziganshin AM, Schmidt T, Scholwin F, et al. Bacteria and archaea involved in anaerobic digestion of distillers grains with solubles. Appl Microbiol Biotechnol, 2011, 89(6): 2039-2052. DOI:10.1007/s00253-010-2981-9 |
| [25] | Oberhardt MA, Zarecki R, Gronow S, et al. Harnessing the landscape of microbial culture media to predict new organism-media pairings. Nat Commun, 2015, 6: 8493. DOI:10.1038/ncomms9493 |
