

北京化工大学 生命科学与技术学院,北京 100029
收稿日期:2020-06-22;接收日期:2020-09-22
基金项目:国家重点研发计划(No. 2018YFA0900200),国家自然科学基金(No. 31870075)资助
作者简介:李正军??博士,北京化工大学教授,博士生导师,主要从事代谢工程与合成生物学领域的教学科研工作,以目标导向的新型细胞工厂构建为主线,针对组合生物合成网络的关键科学问题开展研究,开发出一系列能够利用木糖和乙酸等非常规碳源为底物合成多种化学品的优良底盘细胞。主持国家自然科学基金和国家科技支撑计划等项目10余项,在Metab Eng等期刊发表SCI论文30余篇,申请发明专利13项,获得授权6项.
摘要:海生杆菌属首次于1997年鉴定,迄今包括18个物种,其中10个已完成全基因组序列测定。文中总结了海生杆菌属的菌种特征,并从碳源利用、聚羟基脂肪酸酯代谢和芳香族化合物降解三个方面对基因组测序数据进行了分析。研究发现,海生杆菌属具有完整的糖酵解途径和三羧酸循环,缺乏木糖利用基因。所有海生杆菌属菌种均含有Ⅰ型和Ⅲ型聚羟基脂肪酸酯合成酶的编码基因,表明该菌属可能具有普遍的聚羟基脂肪酸酯合成能力。海生杆菌属含有芳香族化合物的降解途径,苯、苯酚和苯甲酸可由不同的酶催化生成邻苯二酚,再由邻位断裂途径降解为3-酮己二酸,邻苯二酚也可由间位断裂途径降解为丙酮酸和乙酰辅酶A。基因组测序数据分析加深了对海生杆菌属代谢特征的认识,提示该菌属在聚羟基脂肪酸酯合成和海洋芳香族污染物治理方面有一定的应用前景。
关键词:海生杆菌属基因组测序聚羟基脂肪酸酯芳香族化合物
Analysis of the genome sequencing data of the Marinobacterium genus
Mengru Wang, Wei Xi, Zhengjun Li


College of Life Science and Technology, Beijing University of Chemical Technology, Beijing 100029, China
Received: June 22, 2020; Accepted: September 22, 2020
Supported by: National Key Research and Development Program of China (No. 2018YFA0900200), National Natural Science Foundation of China (No. 31870075)
Corresponding author: Zhengjun Li. Tel: +86-10-64421335; Fax: +86-10-64416428; E-mail: lizj@mail.buct.edu.cn.
Abstract: The marine genus Marinobacterium was first identified in 1997, and a total of 18 species have been characterized so far, 10 of which have published whole-genome sequencing data. This article summarizes the characteristics of Marinobacterium genus and analyzes the genome sequencing data related to the carbon source utilization, polyhydroxyalkanoate metabolism, and aromatic compounds degradation. The Marinobacterium species possess the complete glycolysis pathway and tricarboxylic acid cycle, yet lack genes involved in xylose utilization. All strains of the Marinobacterium genus contain the genes encoding for the typeⅠand type Ⅲ polyhydroxyalkanoate synthases, suggesting that the genus may have ability of polyhydroxyalkanoate accumulation. The Marinobacterium species contain the degradation pathways of aromatic compounds. Benzene, phenol and benzoic acid can be degraded into catechol via different enzymes, subsequently catechol is converted to 3-ketoadipate through the ortho-cleavage pathway. Alternatively, catechol can be degraded into pyruvate and acetyl-CoA. The analysis of genome sequencing data of the Marinobacterium genus provides in-depth understanding of the metabolic characteristics, indicating that the genus may have certain applications in the synthesis of polyhydroxyalkanoate and the removal of marine aromatic compounds.
Keywords: Marinobacteriumgenomesequencepolyhydroxyalkanoatearomatic compounds
海洋占据了地球表面的绝大部分,其中蕴含着非常丰富的生物资源。能够在海洋环境中生活的微生物包括细菌、放线菌、蓝细菌、真菌和微藻等多种类群,据估计种类达2亿–10亿种[1]。海洋微生物参与了海洋环境中的物质和能量循环,对维持海洋系统的生态平衡发挥了重要作用。由于海洋环境的多变性、开放性和复杂性,海洋微生物在长期进化过程中形成了适应寡营养、低温、高压和高盐等极端环境的特殊机制,具有很高的生理代谢多样性、遗传多样性和物种多样性。从海洋微生物中筛选结构新、活性优的生理活性物质和新型降解酶类,已经成为相关领域的研究热点[2-3]。开发利用海洋微生物资源,能够为新材料、新能源和新药的生产提供更多研究思路。
海洋微生物常见的种类包括弧菌属Vibrio、假单胞菌属Pseudomonas、螺菌属Spirillum、微球菌属Micrococcus和链霉菌属Streptomyces等[4]。海生杆菌属Marinobacterium最初是González等于1997年鉴定,属于变形菌门、γ变形菌纲、海洋螺菌目、海洋螺菌科。该菌属均为革兰氏阴性细菌,细胞为杆状,大多数菌种的最适生长pH 7–8、NaCl浓度约3%、温度约30 ℃,能够利用糖类、脂肪酸、芳香族化合物和氨基酸等生长[5]。代表菌种是乔治亚海生杆菌Marinobacterium georgiense,能够利用糖类、氨基酸和苯酚、苯甲酸等芳香族化合物以及香豆酸、肉桂酸等木质素相关化合物作为唯一碳源生长[5]。芳香族化合物结构稳定,不易分解,在一定条件下具有致癌、致畸和致突变性。多种微生物具有芳香族化合物降解能力,是去除环境中芳香族污染物最有效、经济和安全的方法[6]。能够代谢芳香族化合物生长的海生杆菌属可能在海洋芳香族污染物的自然消减中发挥作用。
到目前为止,海生杆菌属已经扩展到18个菌种[5, 7-23],部分具有在细胞内合成聚-3-羟基丁酸酯(Poly-3-hydroxybutyrate,PHB)的能力。PHB是发现最早、结构最简单、研究最多的聚羟基脂肪酸酯(Polyhydroxyalkanoate,PHA)。PHA由微生物在生长代谢不平衡条件下在细胞内合成,具有类似塑料的材料学性质,可生物降解,应用前景广泛[24]。PHA的生产菌种主要包括罗氏真氧菌Ralstonia eutropha、嗜水气单胞菌Aeromonas hydrophila、恶臭假单胞菌Pseudomonas putida和盐单胞菌Halomonas等[25],其中盐单胞菌能在高盐条件下进行不灭菌发酵生产PHA,能够显著降低生产成本,正在进行工业化试生产[26]。开发新的PHA生产菌,从而拓宽底物利用范围、提高发酵产量、获得新型PHA材料等,对于促进PHA的产业化应用具有重要意义。
影响微生物适应环境和发挥功能的因素有很多,营养元素中的碳源尤为重要。本文系统总结了海生杆菌属的菌种特征,对已完成基因组测序的10个物种的基因组数据进行了分析,重点研究了碳源代谢、PHA合成与降解、芳香族化合物降解等基因的分布情况。基因组测序数据的分析表明,海生杆菌属普遍具有PHA代谢和芳香族化合物降解的基因,值得进行深入的分子生物学与代谢工程研究,有望在聚羟基脂肪酸酯合成和海洋芳香族污染物治理领域发挥重要作用。
1 材料与方法1.1 数据库和软件海生杆菌属的基因组数据信息来源于NCBI数据库(National Center for Biotechnology Information,https://www.ncbi.nlm.nih.gov/)。系统发育树分析使用软件MEGA (版本7.0)[27],基因组数据的保存和分析等使用软件SnapGene (版本4.3.6)[28]。
1.2 系统发育树分析在NCBI数据库中检索海生杆菌属各菌种的16S rRNA序列和PHA合成酶的氨基酸序列。使用MEGA软件的Neighbor-Joining法[27]对16S rRNA序列和PHA合成酶的氨基酸序列构建相应的系统发育树。
1.3 碳源代谢相关基因分析利用SnapGene软件[28],结合NCBI的Blast功能,分析碳源利用途径中关键酶编码基因在海生杆菌属基因组中的分布情况。具体包括糖酵解途径中的6-磷酸果糖激酶(6-phosphofructokinase)、丙酮酸激酶(Pyruvate kinase),磷酸戊糖途径中的6-磷酸葡萄糖脱氢酶(Glucose-6-phosphate dehydrogenase,G6P dehydrogenase)、6-磷酸葡萄糖酸内酯酶(6-phosphogluconolactonase)、6-磷酸葡萄糖酸脱氢酶(6-phosphogluconate dehydrogenase),Entner-Doudoroff (ED)途径的磷酸葡萄糖酸脱水酶(Phosphogluconate dehydratase)、2-酮-3脱氧-6-磷酸葡萄糖酸醛缩酶(2-dehydro-3-deoxy-phosphogluconate aldolase,KDPG aldolase),乙酸代谢相关的乙酰辅酶A合成酶(Acetate-CoA synthase)、乙酸激酶(Acetate kinase)、磷酸乙酰基转移酶(Phosphate acetyltransferase),以及丙酮酸代谢相关的丙酮酸脱氢酶(Pyruvate dehydrogenase)、丙酮酸氧化酶(Pyruvate oxidase)等。
1.4 聚羟基脂肪酸酯代谢相关基因分析聚羟基脂肪酸酯代谢相关基因的分析方法如1.3所述,分析的酶包括PHA合成酶(PHA synthase)和PHA降解酶(PHA depolymerase)。
1.5 芳香族化合物降解相关基因分析芳香族化合物降解相关基因的分析方法如1.3所述,分析的酶包括催化苯生成邻苯二酚的苯酚羟化酶(Phenol hydroxylase),催化苯甲酸生成邻苯二酚的苯甲酸-1, 2-双加氧酶(Benzoate 1, 2-dioxygenase),催化邻苯二酚生成3-酮己二酸的邻苯二酚1, 2-双加氧酶(Catechol 1, 2-dioxygenase)、黏糠酸环异构酶(Muconate cycloisomerase)、黏糠酸内酯异构酶(Muconolactone delta-isomerase)、3-酮己二酸烯醇内酯酶(3-oxoadipate enol-lactonase),催化邻苯二酚生成丙酮酸和乙酰辅酶A的邻苯二酚2, 3-双加氧酶(Catechol 2, 3-dioxygenase)、2-羟基黏糠酸半醛脱氢酶(2-hydroxymuconic semialdehyde dehydrogenase)、4-草酰巴豆酸互变异构酶(4-oxalocrotonate tautomerase)等。
2 结果与分析2.1 海生杆菌属的特征与基因组测序到目前为止,海生杆菌属共发现并鉴定了18个物种,表型等信息归纳总结如表 1所示,其中已完成基因组测序的菌种有10个。海生杆菌属有16个菌种为好氧菌,2个菌种为兼性厌氧菌。值得注意的是,有8个菌种在鉴定时发现具有在细胞内合成PHB的能力。
表 1 海生杆菌属的表型特征Table 1 Phenotypic characteristics of the Marinobacterium genus
| Strain | Growth range | Relationship with O2 | PHB accumulation | Identification | Genome sequencing | References | ||
| Temperature (℃) | pH | NaCl (%) | ||||||
| M. georgiense | 4-41 (37) | 5.5-9.5 (7.5) | 0.1-11.7 (0.6-2.9) | Aerobic | - | 1997 | 2016 | [5] |
| M. stanieri | 40 | ND | ND | Aerobic | + | 1983 | 2011 | [7, 9] |
| M. jannaschii | ND | ND | ND | Aerobic | - | 1984 | 2014 | [8-9] |
| M. halophilum | 4-37 | 5.3-93 | 3.0-12.0 | Aerobic | ND | 2007 | 2018 | [10] |
| M. litorale | 8-42 (30) | 5.0-12.0 (9) | 1.0-7.5 (3.0-3.5) | Facultatively anaerobic | - | 2007 | 2013 | [11] |
| M. rhizophilum | 5-30 (25) | 6.0-9.0 (7.0) | 1.0-5.0 (3.0) | Aerobic | + | 2008 | 2013 | [12] |
| M. marisflavi | 15-42 (30) | 5.0-11.0(7.0-8.0) | 1.5-7.5 (2.5-3.0) | Aerobic | - | 2009 | ND | [13] |
| M. maritimum | 7-37 (25-28) | 5.5-9.0 (7.5-8.0) | 0.5-7.0 (1.0-2.0) | Aerobic | ND | 2009 | ND | [14] |
| M. sediminicola | 15-42 (35) | 6.0-9.5 (7.0) | 0.5-7.5 (1.0-3.0) | Aerobic | + | 2009 | ND | [15] |
| M. nitratireducens | 15-40 (35) | 5.5-9.5 (7.0-8.0) | 0.5-7.5 (1.0-3.0) | Aerobic | + | 2009 | ND | [15] |
| M. lutimaris | 15-40 (25-30) | 6.0-8.0 (6.5-7.5) | 1.0-10.0(2.0-5.0) | Aerobic | + | 2010 | 2016 | [16] |
| M. coralli | 15-42 (20-40) | ND | 1.0-7.0 | Aerobic | ND | 2011 | ND | [17] |
| M. mangrovicola | 4-42 (28-37) | 5.5-10.0 (6.5-8.0) | 0-18.0(1.0-4.0) | ND | + | 2014 | 2019 | [18] |
| M. aestuariivivens | 10-40 (30) | 6.0 (7.0-8.0) | 0.5-8.0 (0.5-2.0) | Aerobic | ND | 2016 | ND | [19] |
| M. profundum | 4-32 (25-30) | 6.0-9.0 (7.0-7.5) | 1.0-4.0 (3.0) | Aerobic | ND | 2016 | 2016 | [20] |
| M. zhoushanense | 15-43 (37-40) | 5.5-9.5 (6.5-7.5) | 0.25-9.0(1.0-1.5) | Facultatively anaerobic | + | 2016 | ND | [21] |
| M. aestuarii | 4-35 (20-25) | 5.0-9.0 (7, 0-8.0) | 1.0-8.0 (3.0) | Aerobic | ND | 2018 | 2016 | [22] |
| M. boryeongense | 15-45 (25) | 5.5-10.0 (6.0) | 1.0-9.0 (3.0) | Aerobic | + | 2019 | ND | [23] |
| +: positive; -: negative; ND: no data available from the reference. Values in parentheses represent optimal growth conditions. | ||||||||
表选项
在NCBI检索共得到30个完成基因组序列测定的海生杆菌属菌株,其中19个没有确定的菌种归属。10个已完成测序的菌种中,只有M. stanieri包括2个菌株,编号分别为DSM7027和S30。本文选择10个具有确定物种归属的海生杆菌属菌株的基因组序列进行分析,其中M. stanieri选择模式菌株DSM7027,详细信息如表 2所示。10种细菌的基因组的大小差距比较大,最小的基因组为3 653 180 bp (M. halophilum),最大的基因组为5 568 156 bp (M. lutimaris)。海生杆菌属的tRNA种类比较多,有8个菌株的tRNA种数都超过了60种,rRNA和ncRNA种类比较少,都低于10种。基因组的GC含量在(54.9–58.8) mol%之间。
表 2 海生杆菌属的基因组概况Table 2 The genome overview of the Marinobacterium genus
| M. mangrovicola DSM27697 | M. lutimaris DSM22012 | M. rhizophilum DSM18822 | M. stanieri DSM7027 | M. georgiense DSM11526 | M. litorale DSM23545 | M. halophilum DSM17586 | M. profundum PAMC27536 | M. jannaschii DSM6295 | M. aestuarii ST58-10 | |
| Size (bp) | 4 979 440 | 5 568 156 | 5 360 582 | 4 679 482 | 3 922 811 | 4 378 172 | 3 653 180 | 5 637 742 | 5 174 280 | 5 191 608 |
| GC content (%) | 57.1 | 57.4 | 58.5 | 55.6 | 54.9 | 56.4 | 56.0 | 57.2 | 55.2 | 58.8 |
| Genes (total) | 4518 | 5 129 | 4 768 | 4 380 | 3 742 | 4 252 | 3 443 | 5 061 | 4 705 | 4 617 |
| CDSs (total) | 4 443 | 5 047 | 4 691 | 4 296 | 3 673 | 4 179 | 3 365 | 4 968 | 4 629 | 4 512 |
| CDSs (protein) | 4 414 | 5 002 | 4 604 | 4 259 | 3 625 | 4 114 | 3 326 | 4 863 | 4 569 | 4 459 |
| tRNAs | 68 | 69 | 66 | 73 | 56 | 57 | 71 | 82 | 62 | 83 |
| rRNAs | ND | 4 | 6 | 6 | 8 | 9 | 3 | 5 | 8 | 18 |
| ncRNAs | 7 | 9 | 5 | 5 | 5 | 7 | 4 | 6 | 6 | 4 |
表选项
2.2 海生杆菌属的16S rRNA系统发育分析对已经鉴定的所有18个海生杆菌属菌种的模式菌株16S rRNA与同为海洋细菌的Marinobacter属和Neptunomonas属相关菌株进行系统发育树分析,结果如图 1所示。整体来看,海生杆菌属与Neptunomonas属亲缘关系较近,其中M. marisflavi与Neptunomonas属亲缘关系最近,海生杆菌属与Marinobacter属亲缘关系相对较远。海生杆菌属内,M. rhizophilum、M. aestuarii和M. profundum之间的亲缘关系比较近,M. georgiense和M. halophilum之间的亲缘关系比较近。
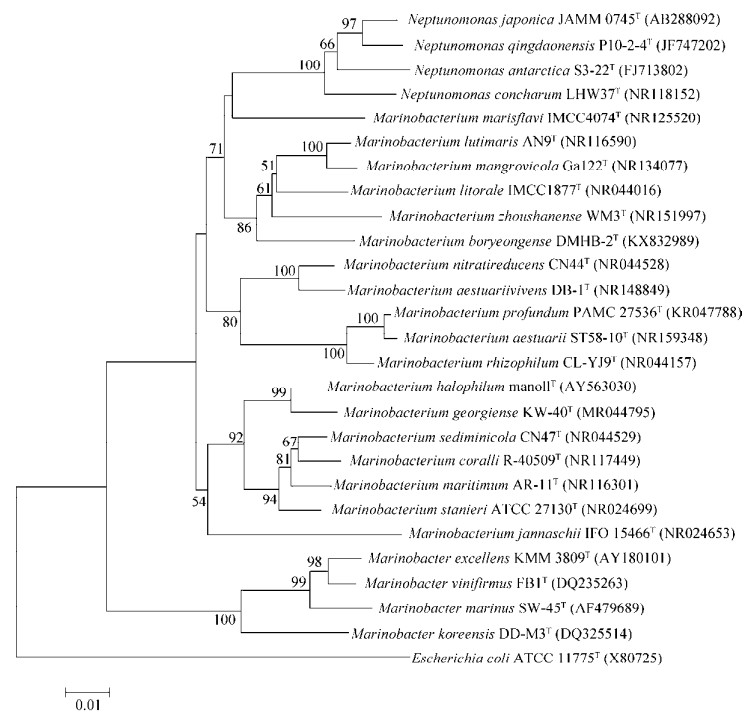 |
| 图 1 海生杆菌属的16S rRNA系统发育树 Fig. 1 Neighbor-joining tree based on 16S rRNA gene sequences showing the phylogenetic relationships of the Marinobacterium genus and some other related taxa. Escherichia coli ATCC 11775T (X80725) was used as the outgroup. The GenBank accession numbers were in the parentheses after bacterial names. Numbers at the branch ends are the bootstrap values based on 1 000 replicates. The scale bar indicates the number of nucleotide substitution per site. |
| 图选项 |
2.3 海生杆菌属的碳源利用途径分析了解碳源利用途径对研究微生物的生长和代谢特征以及应用开发具有重要意义。基于基因组测序数据对海生杆菌属的碳源利用途径进行分析,糖酵解途径、磷酸戊糖途径、ED途径以及乙酸和丙酮酸代谢的部分关键酶基因座位如表 3所示。海生杆菌属中9个菌种含有糖酵解途径的6-磷酸果糖激酶,全部菌种都含有丙酮酸激酶。磷酸戊糖途径和ED途径的关键酶,包括6-磷酸葡萄糖脱氢酶、6-磷酸葡萄糖酸内酯酶、磷酸葡萄糖酸脱水酶、KDPG醛缩酶,在6个菌种中有发现,在M. georgiense、M. halophilum、M. stanieri和M. jannaschii等4个菌种中缺失。10个菌种都没有发现催化6-磷酸葡萄糖酸生成5-磷酸核酮糖的6-磷酸葡萄糖酸脱氢酶,提示它们不能将葡萄糖单独通过磷酸戊糖途径进行分解。乙酸代谢方面,全部菌种都能找到乙酰辅酶A合成酶、乙酸激酶和磷酸乙酰基转移酶,表明海生杆菌属具有利用乙酸的能力,且两条乙酸利用途径同时存在。海生杆菌属都含有丙酮酸脱氢酶和完整的柠檬酸循环。五碳糖利用方面,没有发现与木糖代谢相关的木糖异构酶(Xylose isomerase)和木糖脱氢酶(Xylose dehydrogenase)等,提示海生杆菌属不具有发酵木糖的能力。
表 3 海生杆菌属中碳源代谢相关基因的基因座位Table 3 The locus tags of enzymes rel ated to carbon metabolism in the Marinobacterium genus
| Enzyme name | M. georgiense | M. stanieri | M. jannaschii | M. halophilum | M. litorale | M. rhizophilum | M. lutimaris | M. mangrovicola | M. projundum | M. aestuarii |
| DSM11526 | DSM7027 | DSM6295 | DSM17586 | DSM23545 | DSM18822 | DSM22012 | DSM27697 | PAMC27536 | ST58-10 | |
| 6-phosphofructokinase | RSI6265 | RS19815 | RS0121735 | RS09325 | RS0118635 | RS0108970 | RS22575 | RSI9675 | NA | RS19875 |
| Pyruvate kinase | RS09215 | RSI8250 | RS0122890 | RS07430 | RS0106800 | RS0104495 | RS18890 | RS15015 | RS14810 | RS12415 |
| G6P dehydrogenase | NA | NA | NA | NA | RS0115305 | RS0111030 | RS00095 | RS21800 | RS16710 | RSI1110 |
| 6-phosphogluconolactonase | NA | NA | NA | NA | RS0115300 | RS0111035 | RSI1295 | RSI1725 | RSI6705 | RSI1105 |
| Phosphogluconate | NA | NA | NA | NA | RS0115285 | RS0110140 | RSI1305 | RS11715 | RSI3685 | RSI1775 |
| Dehydratase | ||||||||||
| KDPG aldolase | NA | NA | NA | NA | RS0115290 | RS0111040 | RSI1300 | RSI1720 | RSI6700 | RSI1100 |
| Acetate-CoA synthase | RS02360 | RS20015 | RS0112200 | RSI2750 | RS0103075 | RS0112780 | RS13510 | RS09460 | RS04525 | RS04115 |
| Acetate kinase | RS07295 | RSI1655 | RS24355 | RS04290 | RS0104140 | RS0115065 | RSI6970 | RS20745 | RS07240 | RS17275 |
| Phosphate acetyltransferase | RS07300 | RSI1650 | RS0102680 | RS04285 | RS0104145 | RS0115070 | RSI6965 | RS20740 | RS07245 | RS17270 |
| Pyruvate dehydrogenase | RS07985 | RS01365 | RS0121835 | RS03690 | RS0117295 | RS0120165 | RSI6425 | RS20180 | RS23800 | RS05075 |
| RS07990 | RS01360 | RS0121830 | RS03685 | RS0117290 | RS0120170 | RSI6420 | RS20175 | RS23795 | RS05080 | |
| RS07995 | RS01355 | RS0121825 | RS03680 | RS0117285 | RS0120175 | RS16415 | RS20170 | RS23790 | RS05085 | |
| The prefix part of the locus tag indicating the strain is omitted. NA: no sequence available through the genome analysis. | ||||||||||
表选项
2.4 海生杆菌属的聚羟基脂肪酸酯代谢分析根据海生杆菌属的菌种鉴定文献,8个菌种具有PHB合成能力(表 1)。微生物可以利用糖类和脂肪酸等不同碳源经由多种代谢途径形成羟基酯酰辅酶A单体,再由PHA合成酶催化合成聚酯。PHA合成酶的性质决定了聚合物的单体组成、比例和分子量等,是PHA合成途径的最关键酶。PHA合成酶根据其结构、亚基组成和底物特异性等可分为4种类型。对来源于假单胞菌的PHA合成酶进行定向进化,实现了2-羟基酯酰辅酶A单体的聚合,合成出乳酸和3-羟基丁酸共聚酯、乳酸和乙醇酸共聚酯等[29-30]。PHA分解再利用的第一步是由PHA降解酶催化聚合物酯键水解释放羟基脂肪酸单体。本文重点关注在天然能够合成聚酯的菌种中普遍存在的PHA合成酶和PHA降解酶,对海生杆菌属基因组测序数据进行分析,相关基因座位如表 4所示。海生杆菌属的全部菌种都含有2–3个PHA合成酶,对其与4种类型PHA合成酶的代表进行系统发育树分析(图 2),结果发现9个菌种都分别含有3个PHA合成酶,其中2个为Ⅰ型,1个为Ⅲ型。Ⅲ型合成酶包含2个亚基,编码基因形成一个操纵子转录,并且毗邻PHA合成阻遏蛋白。海生杆菌属的2个Ⅰ型PHA合成酶之间的序列一致性较低,系统发育树中2个Ⅰ型PHA合成酶PhaC1和PhaC2位于相对独立的分支中,提示其可能属于旁系同源。海生杆菌属的PHA合成酶组成和分布情况与同为海洋细菌的Neptunomonas属十分类似,与罗氏真氧菌、嗜水气单胞菌和恶臭假单胞菌等传统PHA生产菌有很大差别[31]。此外,海生杆菌属普遍存在PHA降解酶基因,表明其具有重新分解利用细胞内积累的聚羟基脂肪酸酯的能力,不过有3个菌种的PHA降解酶基因没有找到,可能与基因组测序数据的质量有关。
表 4 海生杆菌属中聚羟基脂肪酸酯代谢相关基因的基因座位Table 4 The locus tags of genes associated with polyhydroxyalkanoate metabolism in the Marinobacteriumn genus
| Enzyme name | M. georgiense DSM11526 | M. stanieri DSM7027 | M. jannaschii DSM6295 | M. halophilum DSM17586 | M. litorale DSM23545 | M. rhizophilum DSM18822 | M. lutimaris DSM22012 | M. mangrovicola DSM27697 | M. profundum PAMC27536 | M. aestuarii ST58-10 |
| PHA synthase PhaCl | RS03545 | RS05215 | RS0119885 | RS05365 | RS0108580 | RS0105515 | RS02135 | RS07845 | RS07445 | RS13030 |
| PHA synthase PhaC2 | RS03730 | RS05405 | NA | RS04860 | RS0108390 | RS0116830 | RS01960 | RS08020 | RS08540 | RS12595 |
| PHA synthase subunit PhaC | RS03395 | RS05055 | RS0119725 | RS05500 | RS0108735 | RS0105665 | RS02285 | RS07695 | RS07595 | RS12880 |
| PHA synthase subunit PhaE | RS03390 | RS05050 | RS0119720 | RS05505 | RS0108740 | RS0105670 | RS02290 | RS07690 | RS07600 | RS12875 |
| PHA synthesis repressor PhaR | RS03400 | RS05060 | RS0119730 | RS05495 | RS0108730 | RS24915 | RS02280 | RS07700 | RS07590 | RS12885 |
| PHA depolymerase PhaZ | NA | NA | RS0119360 | NA | RS0109630 | RS0108745 | RSI1455 | RSI 1555 | RS09845 | RS10290 |
| The prefix part of the locus tag indicating the strain is omitted. NA: no sequence available through the genome analysis. | ||||||||||
表选项
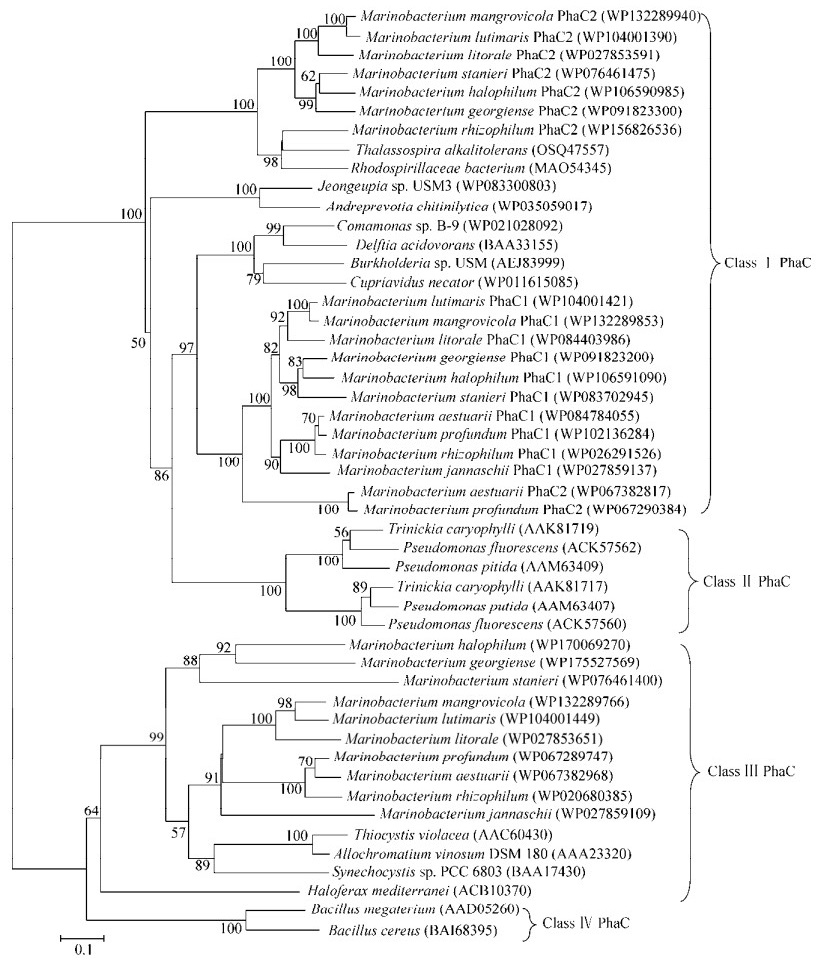 |
| 图 2 海生杆菌属PHA合成酶及其他代表性PHA合成酶的发育树 Fig. 2 Phylogenetic tree of the PHA synthases from the Marinobacterium genus and related taxa using amino acid sequence. The GenBank accession numbers were in the parentheses after bacterial names. Numbers at the branch ends are the bootstrap values based on 1 000 replicates. The scale bar indicates the number of amino acid substitution per site. |
| 图选项 |
2.5 海生杆菌属的芳香族化合物代谢分析作为海生杆菌属的代表菌种,乔治亚海生杆菌能够利用苯酚等芳香族化合物作为唯一碳源生长[5]。基于基因组测序数据对海生杆菌属的芳香族化合物降解途径进行分析,发现该菌属普遍具有苯、苯酚和苯甲酸的降解途径(表 5)。芳香族化合物经过以邻苯二酚为中间代谢物的途径进行降解,相关基因以基因簇的形式存在。苯和苯酚由苯酚羟化酶催化生成邻苯二酚;苯甲酸由苯甲酸-1, 2-双加氧酶催化生成邻苯二酚。邻苯二酚经由邻位断裂途径降解为3-酮己二酸,或者由间位断裂途径降解为丙酮酸和乙酰辅酶A。邻位断裂途径包括4个酶:邻苯二酚1, 2-双加氧酶、黏糠酸环异构酶、黏糠酸内酯异构酶和3-酮己二酸烯醇内酯酶;3-酮己二酸可以进一步与辅酶A缩合,再分解成琥珀酸和乙酰辅酶A,从而进入三羧酸循环被彻底氧化。间位断裂途径包括邻苯二酚2, 3-双加氧酶、2-羟基黏糠酸半醛脱氢酶、4-草酰巴豆酸互变异构酶等7个酶,将邻苯二酚降解为丙酮酸和乙酰辅酶A,再进入三羧酸循环氧化。据报道,同为海洋细菌的Neptunomonas属也有芳香族化合物的降解途径,邻苯二酚由间位断裂途径降解[31]。在某些假单胞菌中,可以同时具有邻位断裂和间位断裂两条途径,在高浓度苯甲酸底物存在时,两条途径能够同时发挥降解作用[32]。海生杆菌属具体的芳香族化合物降解能力需要进一步实验研究。
表 5 海生杆菌属中芳香族化合物降解相关基因的基因座位Table 5 The locus tags of enzymes related to degradation of aromatic compounds in the Marinobacterium genus
| Enzyme name | M. georgiense DSM11526 | M. stanieri DSM7027 | M. jannaschii DSM6295 | M. halophilum DSM17586 | M. Utorale DSM23545 | M. rhizophilum DSM18822 | M. profundum PAMC27536 | M. aestuarii ST58-10 |
| Phenol hydroxylase | RS06070 | RSI1270 | RS0100825 | RSI1105 | RS0106405 | NA | RS09750 | RS10400 |
| RS06075 | RSI1265 | RS0100830 | RSI1100 | RS0106400 | NA | RS09745 | RS10405 | |
| RS06080 | RSI1260 | RS0100835 | RSI1095 | RS0106395 | NA | RS09740 | RS10410 | |
| RS06085 | RSI1255 | RS0100840 | RSI1090 | RS0106390 | NA | RS09735 | RS10415 | |
| RS06090 | RSI1250 | RS0100845 | RSI1085 | RS0106385 | NA | RS09730 | RSI0420 | |
| RS06095 | RSI1245 | RS0100850 | RSI1080 | RS0106380 | NA | RS09725 | RS10425 | |
| Benzoate 1, 2-dioxygenase | RSI8325 | RS11410 | NA | RSI1205 | RS0101480 | RS0106495 | RS09635 | RSI0505 |
| RSI8330 | RSI1405 | NA | RSI1200 | RS0101485 | RS0106500 | RS09630 | RS10510 | |
| RSI8335 | RSI1400 | NA | RSI1195 | RS0101490 | RS0106505 | RS09625 | RS10515 | |
| RSI8340 | RSI1395 | NA | RSI1190 | RS0101495 | RS0106510 | RS09620 | RSI0520 | |
| Catechol 1, 2-dioxygenase | NA | NA | NA | NA | NA | RS0106490 | RS11360 | RS13505 |
| Muconolactone D-isomerase | NA | NA | NA | NA | NA | RS0106485 | RSI1365 | RS13510 |
| Muconate cycloisomerase | NA | NA | NA | NA | NA | RS0106480 | RSI1370 | RS13515 |
| 3-oxoadipate enol-lactonase | NA | NA | NA | NA | NA | RS0121670 | RS04860 | RS03785 |
| Catechol 2, 3-dioxygenase | RS01380 | RSI1325 | RS0100860 | RSI1070 | RS0106365 | NA | RS20120 | NA |
| 2-hydroxymuconic semialdehyde dehydrogenase | RS01345 | RSI1375 | RS0100865 | RSI1175 | RS0106320 | NA | RS20140 | NA |
| 4-oxalocrotonate tautomerase | RS01330 | RSI1350 | RS0100890 | RS11150 | RS0106290 | NA | RS20165 | NA |
| 2-oxo-3-hexenedioate decarboxylase | RS01335 | RSI1355 | RS0100885 | RS11155 | RS0106295 | NA | RS20160 | NA |
| 2-oxopent-4-enoate hydratase | RS01340 | RSI1370 | RS0100870 | RSI1170 | RS0106310 | NA | RS20145 | NA |
| 4-hydroxy-2-oxovalerate aldolase | RS01320 | RSI1360 | RS0100880 | RSI1160 | RS0106300 | NA | RS20155 | NA |
| Acetaldehyde dehydrogenase | RS01325 | RSI1365 | RS0100875 | RSI1165 | RS0106305 | NA | RS20150 | NA |
| The prefix part of the locus tag indicating the strain is omitted. NA: no sequence available through the genome analysis. | ||||||||
表选项
3 结论本文总结了海生杆菌属已鉴定的18个菌种的特征,并对其中10个完成基因组测序的数据进行了分析。研究结果发现,海生杆菌属普遍含有聚羟基脂肪酸酯合成酶和降解酶基因,表明该菌属可能具有聚羟基脂肪酸酯合成能力,后续选择合适的菌株建立遗传操作方法和代谢工程改造,有望开发能够在海洋高盐条件下生产聚羟基脂肪酸酯的新型底盘细胞。另外,基因组测序数据分析还表明,海生杆菌属具有降解芳香族化合物的相关酶类,含有将苯、苯酚和苯甲酸等经邻苯二酚中间体降解的代谢途径,具有在海洋环境中降解芳香族化合物的功能。本研究加深了对海生杆菌属基因组特征的认识,为该菌属在聚羟基脂肪酸酯合成和海洋芳香族化合物污染治理等方面的应用提供了研究基础。
参考文献
| [1] | Zhang S, Zhang CS, Tian XP, et al. The study of diversities of marine microbes in China. Bull Chin Acad Sci, 2010, 25(6): 651-658 (in Chinese). 张偲, 张长生, 田新朋, 等. 中国海洋微生物多样性研究. 中国科学院院刊, 2010, 25(6): 651-658. DOI:10.3969/j.issn.1000-3045.2010.06.011 |
| [2] | Wen X, Zhou SL, Yang XJ, et al. Research progress on polysaccharide-degrading enzymes from marine microorganism. Biotechnol Bull, 2016, 32(11): 38-46 (in Chinese). 文霞, 周少璐, 杨秀茳, 等. 海洋微生物多糖降解酶的研究进展. 生物技术通报, 2016, 32(11): 38-46. |
| [3] | Fan ML, Yu BY, Gu JF. Research advances of the antibiotics substances of marine microorganisms. World Notes Antibiot, 2014, 35(4): 145-148 (in Chinese). 樊梦霖, 余伯阳, 顾觉奋. 海洋微生物抗菌活性物质研究进展. 国外医药:抗生素分册, 2014, 35(4): 145-148. |
| [4] | Ding YJ, Ding MD, Zhang JD, et al. Quorum sensing in marine microorganisms. Bull Sci Technol, 2019, 35(6): 1-6 (in Chinese). 丁雅娟, 丁蒙丹, 张佳娣, 等. 海洋微生物中的群体感应. 科技通报, 2019, 35(6): 1-6. |
| [5] | González JM, Mayer F, Moran MA, et al. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Evol Microbiol, 1997, 47(2): 369-376. |
| [6] | Meynet P, Head IM, Werner D, et al. Re-evaluation of dioxygenase gene phylogeny for the development and validation of a quantitative assay for environmental aromatic hydrocarbon degraders. FEMS Microbiol Ecol, 2015, 91(6): fiv049. |
| [7] | Baumann P, Bowditch RD, Baumann L, et al. Taxonomy of marine Pseudomonas species: P. stanieri sp nov.; P. perfectomarina sp. nov., nom. rev.; P. nautica: and P. doudorofii. Int J Syst Evol Microbiol, 1983, 33(4): 857-865. |
| [8] | Bowditch RD, Baumann L, Baumann P. Description of Oceanospirillum kriegii sp. nov. and O. jannaschii sp. nov. and assignment of two species of Alteromonas to this genus as O. commune comb. nov. and O. vagum comb. nov. Curr Microbiol, 1984, 10(4): 221-229. DOI:10.1007/BF01627259 |
| [9] | Satomi M, Kimura B, Hamada T, et al. Phylogenetic study of the genus Oceanospirillum based on 16S rRNA and gyrB genes: emended description of the genus Oceanospirillum, description of Pseudospirillum gen. nov., Oceanobacter gen. nov. and Terasakiella gen. nov. and transfer of Oceanospirillum jannaschii and Pseudomonas stanieri to Marinobacterium as Marinobacterium jannaschii comb. nov. and Marinobacterium stanieri comb. nov. Int J Syst Evol Microbiol, 20002, 52(3): 739-747. |
| [10] | Chang HW, Nam YD, Kwon HY, et al. Marinobacterium halophilum sp. nov., a marine bacterium isolated from the Yellow Sea. Int J Syst Evol Microbiol, 2007, 57(1): 77-80. DOI:10.1099/ijs.0.64505-0 |
| [11] | Kim H, Choo YJ, Song J, et al. Marinobacterium litorale sp. nov. in the order Oceanospirillales. Int J Syst Evol Microbiol, 2007, 57(7): 1659-1662. DOI:10.1099/ijs.0.64892-0 |
| [12] | Kim YG, Jin YA, Hwang CY, et al. Marinobacterium rhizophilum sp. nov., isolated from the rhizosphere of the coastal tidal-flat plant Suaeda japonica. Int J Syst Evol Microbiol, 2008, 58(1): 164-167. DOI:10.1099/ijs.0.65176-0 |
| [13] | Kim H, Oh HM, Yang SJ, et al. Marinobacterium maris?avi sp. nov., isolated from a costal seawater. Curr Microbiol, 2009, 58(5): 511-515. DOI:10.1007/s00284-009-9355-5 |
| [14] | Kim SJ, Park SJ, Yoon DN, et al. Marinobacterium maritimum sp. nov., a marine bacterium isolated from Arctic sediment. Int J Syst Evol Microbiol, 2009, 59(12): 3030-3034. DOI:10.1099/ijs.0.009134-0 |
| [15] | Huo YY, Xu XW, Cao Y, et al. Marinobacterium nitratireducens sp. nov. and Marinobacterium sediminicola sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol, 2009, 59(5): 1173-1178. DOI:10.1099/ijs.0.005751-0 |
| [16] | Kim JM, Lee SH, Jung JY, et al. Marinobacterium lutimaris sp. nov., isolated from a tidal flat. Int J Syst Evol Microbiol, 2010, 60(8): 1828-1831. DOI:10.1099/ijs.0.016246-0 |
| [17] | Chimetto LA, Cleenwerck I, Brocchi M, et al. Marinobacterium coralli sp. nov., isolated from mucus of coral (Mussismilia hispida). Int J Syst Evol Microbiol, 2011, 61(1): 60-64. DOI:10.1099/ijs.0.021105-0 |
| [18] | Alfaro-Espinoza G, Ullrich MS. Marinobacterium mangrovicola sp. nov., a marine nitrogen-fixing bacterium isolated from mangrove roots of Rhizophora mangle. Int J Syst Evol Microbiol, 2014, 64(12): 3988-3993. |
| [19] | Park S, Jung YT, Kim S, et al. Marinobacterium aestuariivivens sp. nov., isolated from a tidal ?at. Int J Syst Evol Microbiol, 2016, 66(4): 1718-1723. DOI:10.1099/ijsem.0.000927 |
| [20] | Hwang CY, Yoon SJ, Lee I, et al. Marinobacterium profundum sp. nov., a marine bacterium from deep-sea sediment. Int J Syst Evol Microbiol, 2016, 66(3): 1561-1566. DOI:10.1099/ijsem.0.000918 |
| [21] | Han SB, Wang RJ, Yu XY, et al. Marinobacterium zhoushanense sp. nov., isolated from surface seawater. Int J Syst Evol Microbiol, 2016, 66(9): 3437-3442. DOI:10.1099/ijsem.0.001213 |
| [22] | Bae SS, Jung J, Chung D, et al. Marinobacterium aestuarii sp. nov., a benzene-degrading marine bacterium isolated from estuary sediment. Int J Syst Evol Microbiol, 2018, 68(2): 651-656. DOI:10.1099/ijsem.0.002561 |
| [23] | Kang JY, Kim MJ, Chun J, et al. Marinobacterium boryeongense sp. nov., isolated from seawater. Int J Syst Evol Microbiol, 2019, 69(2): 493-497. DOI:10.1099/ijsem.0.003184 |
| [24] | Che XM, Situ W, Yu LS, et al. Application perspectives of polyhydroxyalkanoates. Chin J Biotech, 2018, 34(10): 1531-1542 (in Chinese). 车雪梅, 司徒卫, 余柳松, 等. 聚羟基脂肪酸酯的应用展望. 生物工程学报, 2018, 34(10): 1531-1542. |
| [25] | Li ZJ, Wei XX, Chen GQ. Microbial cell factories for production of polyhydroxyalkanoates. Chin J Biotech, 2010, 26(10): 1426-1435 (in Chinese). 李正军, 魏晓星, 陈国强. 生产聚羟基脂肪酸酯的微生物细胞工厂. 生物工程学报, 2010, 26(10): 1426-1435. |
| [26] | Zhang MY, Li YH, Zhan YL, et al. Research advances in biosynthesis of polyhydroxyalkanoates (PHAs) by halophilic bacteria. Biotechnol Bull, 2019, 35(6): 172-177 (in Chinese). 张梦颖, 李雅慧, 詹元龙, 等. 嗜盐菌生物合成聚羟基脂肪酸酯(PHAs)的研究进展. 生物技术通报, 2019, 35(6): 172-177. |
| [27] | Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016, 33(7): 1870-1874. DOI:10.1093/molbev/msw054 |
| [28] | Daigneault B, Vilarino M, Rajput S, et al. 79 CRISPR gene editing in bovine zygotes-mutation confirmation by integration of protein expression and DNA sequencing analyses. Reprod Fert Dev, 2019, 31(1): 165. |
| [29] | Choi SY, Park SJ, Kim WJ, et al. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat Biotechnol, 2016, 34(4): 435-440. |
| [30] | Taguchi S, Yamada M, Matsumoto K, et al. A microbial factory for lactate-based polyestersusing a lactate-polymerizing enzyme. Proc Natl Acad Sci USA, 2008, 105(45): 17323-17327. DOI:10.1073/pnas.0805653105 |
| [31] | Pu N, Li W, Li ZJ. Complete genome sequence of Neptunomonas concharum JCM17730T: an acetate assimilating bacterium isolated from a dead ark clam. Mar Genom, 2020, 53: 100754. DOI:10.1016/j.margen.2020.100754 |
| [32] | Cao B, Geng AL, Loh KC. Induction of ortho- and meta-cleavage pathways in Pseudomonas in biodegradation of high benzoate concentration: MS identification of catabolic enzymes. Appl Microbiol Biotechnol, 2008, 81(1): 99-107. |
