
 , 康振1,2
, 康振1,2

1 江南大学 工业生物技术教育部重点实验室,江苏 无锡 214122
2 江南大学 食品安全与营养协同创新中心,江苏 无锡 214122
基金项目:国家自然科学基金(No. 31670092),江苏省科技攻关项目(No. BE2014607),****与创新团队发展计划(No. IRT_15R26),江苏省自然科学基金(No. BK20141107) 资助
摘要: 转录终止子作为一种位于终止密码子后的调控信号,负责终止DNA的转录和RNA的释放。文中首次改造并分析了来源于噬菌体的λto终止子的发卡结构与富含尿嘧啶的序列对枯草芽孢杆菌168中基因转录终止效率以及mRNA稳定性的影响。结果表明,相对于野生型的λt0终止子,突变体M3、M11和M12表现出了更高的转录终止效率,突变体M3、M4和M11更有利于上游绿色荧光蛋白mRNA的稳定。另外,我们发现插入RNase作用位点同样提高了mRNA的稳定性。研究结果表明终止子中的发卡环对转录终止不是必需的,同时,结果也证明了转录终止子可以作为一种潜在的工具用于提高枯草芽孢杆菌中mRNA的稳定性以及相应酶的表达。
关键词: 合成生物学 转录终止子 λto终止子 mRNA稳定性 枯草芽孢杆菌
Engineering and characterization of new intrinsic transcriptional terminators in Bacillus subtilis 168
Jean Paul Sinumvayo1, Sen Yang1, Jian Chen1,2, Guocheng Du1,2

 , Zhen Kang1,2
, Zhen Kang1,2

1 The Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu, China;
2 Synergetic Innovation Center of Food Safety and Nutrition, Jiangnan University, Wuxi 214122, Jiangsu, China
Received: December 16, 2016; Accepted: February 8, 2017
Supported by:National Natural Science Foundation of China (No. 31670092), Key Technologies R&D Program of Jiangsu Province, China (No. BE2014607), Program for Changjiang Scholars and Innovative Research Team in University (No. IRT_15R26), Natural Science Foundation of Jiangsu Province (No. BK20141107)
Corresponding author:Guocheng Du. Tel: +86-510-85918307; Fax: +86-510-85918309; E-mail: gcdu@jiangnan.edu.cn
Zhen Kang. Tel: +86-510-85918307; E-mail: zkang@jiangnan.edu.cn
Abstract: Terminators as regulatory signals are typically placed behind the last coding sequence to block the transcription of DNA to RNA and release the transcript. In the present study, the hairpin and the U-rich sequence of the bacteriophage λto terminator were first modified to investigate their effects on termination efficiency and mRNA stability in Bacillus subtilis 168. Compared with the native λto terminator, the terminator variants M3, M11 and M12 showed higher termination efficiency values. Moreover, the variantsM3, M4 and M11 showed significant positive effects on the mRNA stability of the upstream gfp gene. Additionally, insertion of RNase site also increased the mRNA stability. The results of this study suggested that the composition of the hairpin loop is not required for effective intrinsic termination in B. subtilis. Our results also showed that the terminator could also be used as a potential tool for increasing mRNA stability and the corresponding enzyme production in B. subtilis.
Key words: synthetic biology transcription terminator λto terminator mRNA stability Bacillus subtilis
IntroductionTerminators are usually located at the end of a gene or an operon where they terminate the transcription of DNA to RNA and release the transcript[1]. Terminators fall into two categories: 1) rho-independent (or intrinsic) terminators and 2) rho-dependent terminators[2-3]. A majority of previous studies focused on intrinsic terminators, due to their function in the dissociation of transcription complexes without the assistance of auxiliary factors. In bacteria, the intrinsic terminator contains a stable GC-rich hairpin stem loop and a U-stretch that required for disturbing the stable transcription elongation complexes[3-5]. The transcription termination process involves in dissociation of RNA polymerase and separation of the RNA:DNA hybrid [6].
For a long time, terminators have not received enough attention as genetic regulators [2] and most studies have focused on prediction and identification alone[2, 7], as well as the mutation and evaluation of the effects of the GC-rich hairpin and the poly(U) sequence[4, 8-12]. Recently, terminators have been demonstrated to be highly important not only on downstream gene protection against transcript read-through but also for its positive effect on the stabilization of upstream mRNA transcripts [8, 13]. In this regard, Chen et al. built a library of 582 natural and synthetic terminators. After calculation and comparison, they found that in addition to receiving the greatest contribution from the U-tract, the base content of the hairpin stem also correlated with the terminator strength. Specially, they found that strong terminators usually have higher GC content at the bottom of the hairpin stem. In contrast, no or weak correlations were observed between the hairpin stability and its length [14]. Similarly, Cambray et al. characterized 61 natural and synthetic terminators and attested their roles in the quantitative modeling of transcription termination for the optimization of synthetic genetic systems[8]. More recently, Li et al. quantified the relationship between terminator position and terminator efficiency and provided a simple method for fine tuning termination efficiency without changing terminator sequences[11]. In addition to these fundamental studies, many studies have extended the usability of terminators in the construction of genetic systems (such as Logic gates [15-16]and genetic bandpass filter[17-18]), pathway optimization[19]and enzyme expression[9, 13, 20], suggesting the potential powerful applications of terminators in synthetic biology.
To date, most studies have concentrated on E. coli strains and only a small fraction of literatures are focused on other microorganisms[21], such as Bacillus subtilis[1, 22-23]. In fact, the generally recognized safe strain B. subtilis has been widely used as a powerful cell factory for multiple applications, including enzyme production, pathway engineering and synthetic biology[24-27]. Hence, engineering and characterization of rho-independent terminators in B. subtilis is of great significance. In the present study, the bacteriophage λto terminator was engineered and investigated with an experimental system using B. subtilis. The results would have important applications in synthetic biology.
1 Materials and methods1.1 Bacterial strains and plasmidsThe strains, plasmids and primers used in this study are listed in Table 1 and Table 2, respectively. E. coli strain JM109 (e14? (mcrA), endA1, recA1, hsdR17 (rk?, mk+), (lac-proAB) lacIqZM15, relA1) used for plasmid construction and molecular manipulation of all genetic parts and E. coli strain TOP10 (F– mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ (ara-leu) 7697 galE15 galK16 rpsL (StrR) endA1 λ), were purchased from Invitrogen. The pUC57-simple (Ampr, cloning vector in E. coli, isolated from E. coli strain DH5α, MCS) that harbored the rfp gene was obtained from Takara. To perform the assay on the genetic system in this study, samples of B. subtilis 168 (trpC2) obtained from BGSC and pP43NMK (Ampr, Kanr, shuttle expression-secretion vector) were used as expression host and vector, respectively.
Table 1 Strains and plasmids used in this study
| Names | Description | Source |
| Strains E. coli JM109 | e14?(mcrA), endA1, recA1, hsdR17 (rk?, mk+), (lac-proAB) lacIqZM15, relA1 | Invitrogen |
| TOP10 | F–mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ- | Invitrogen |
| B. subtilis 168 | trpC2 | BGSC |
| B. subtilis gfp-λto-rfp | 168 derivate, harboring plasmid pP43NMK-gfp-λto-rfp | This work |
| B. subtilis gfp-rfp | 168 derivate, harboring plasmid pP43NMK-gfp-rfp | This work |
| B. subtilis gfp-RNase-λto-RNase-strong hairpin-rfp | 168 derivate, harboring plasmid pP43NMK-gfp-RNase-λto-RNase-strong hairpin-rfp | This work |
| B. subtilis-gfp | 168 derivate, harboring plasmid pP43NMK-gfp | This work |
| B. subtilis-M1 | 168 derivate, harboring plasmid pP43NMK-M1 | This work |
| B. subtilis-M2 | 168 derivate, harboring plasmid pP43NMK-M2 | This work |
| B. subtilis-M3 | 168 derivate, harboring plasmid pP43NMK-M3 | This work |
| B. subtilis-M4 | 168 derivate, harboring plasmid pP43NMK-M4 | This work |
| B. subtilis-M5 | 168 derivate, harboring plasmid pP43NMK-M5 | This work |
| B. subtilis-M6 | 168 derivate, harboring plasmid pP43NMK-M6 | This work |
| B. subtilis-M7 | 168 derivate, harboring plasmid pP43NMK-M7 | This work |
| B. subtilis-M8 | 168 derivate, harboring plasmid pP43NMK-M8 | This work |
| B. subtilis-M9 | 168 derivate, harboring plasmid pP43NMK-M9 | This work |
| B. subtilis-M10 | 168 derivate, harboring plasmid pP43NMK-M10 | This work |
| B. subtilis-M11 | 168 derivate, harboring plasmid pP43NMK-M11 | This work |
| Plasmids | ||
| pP43NMK | Ampr, Kanr, Shuttle expression-secretion vector | [28] |
| PUC57-simple | Ampr, cloning vector in E. coli, isolated from E. coli strain DH5α, MCS | TaKaRa |
| pP43NMK-gfp-λto-rfp | pP43NMK derivative with gene gfp downstream P43 and followed by λto terminator upstream rfp gene | This work |
| pP43NMK-gfp-rfp | pP43NMK derivative with gfp gene following P43 and followed by rfp gene | This work |
| pP43NMK-gfp-RNase-λto-RNase-strong hairpin-rfp | pP43NMK derivative with gfp gene following P43 and followed by insulated λtoterminator by RNase E site, strong hairpin and then gfp gene | This work |
| pP43NMK-gfp | pP43NMK derivative with gfp gene following P43 | This work |
| pP43NMK-M1 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in T-tail by A to T and followed by upstream rfp gene | This work |
| pP43NMK-M2 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the loop by UGCC to TTCG and followed by upstream rfp gene | This work |
| pP43NMK-M3 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the loop by UGCC to TTCG, A to T in T-tail and followed by upstream rfp gene | This work |
| pP43NMK-M4 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the loop by UGCC to GAAA and followed by upstream rfp gene | This work |
| pP43NMK-M5 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the loop by UGCC to GAAA, A to T in T-tail and followed by upstream rfp gene | This work |
| pP43NMK-M6 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in stem by T to C and followed by upstream rfp gene | This work |
| pP43NMK-M7 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the stem by T to C and T to C closer the loop+ GAAA and followed by upstream rfp gene | This work |
| pP43NMK-M8 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the stem by T to C and T to C closer the loop+ TTCG and followed by upstream rfp gene | This work |
| pP43NMK-M9 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the stem by T to C and T to C closer the loop+ GAAC and followed by upstream rfp gene | This work |
| pP43NMK-M10 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the stem by T to C and T to C closer the loop+ AAATC and followed by upstream rfp gene | This work |
| pP43NMK-M11 | pP43NMK derivative with gfp gene downstream P43 and followed by λto terminator modified in the stem by T to C and T to C closer the loop+ AAAA and followed by upstream rfp gene | This work |
表选项
Table 2 Primers used in this study
| Primer name | Primer sequence (5′–3′) |
| GFP-F | GGGGTACCCTCGAGAAAGGAGGTGAAATGTACACATG |
| GFP-R | AACTGCAGACTAGTTCTAGAAGTACTTTATTTGTATAGTTCATCCATGCC |
| RFP-F | ATAAGAATGCGGCCGCAAGAGAGGAATGTACACATGGCCTCCTCCGAGAAC |
| RFP-R | ATAAGAATGCGGCCGCTGCATAAAAAACGCCCGGCGGCAACCGAGCGTTCT |
| λto-F | CGAGCTCTGATTAATTAATTCAGAACGCTCG |
| λto-R | GGACTAGTTTACAGGAACAGGTGGTGGCGGCC |
| M1-F | ATAAGAATGCGGCCGCTGCAAAAAAAACGCCCGGCGGCAACCGAGCGTTCT |
| M2-F | ATAAGAATGCGGCCGCTGCATAAAAAACGCCCGGCCGAAACCGAGCGTTCT |
| M3-F | ATAAGAATGCGGCCGCTGCAAAAAAAACGCCCGGCCGAAACCGAGCGTTCT |
| M4-F | ATAAGAATGCGGCCGCTGCAAAAAAAACGCCCGGCTTTCACCGAGCGTTCT |
| M5-F | ATAAGAATGCGGCCGCTGCATAAAAAACGCCCGGCTTTCACCGAGCGTTCT |
| M6-F | ATAAGAATGCGGCCGCTGCATAAAAAACGCCCGGCGGCAACCGGGCGTTCT |
| M7-F | CGAGCTCTGATTAATTAATTCAGAACGCCCGGCGAAAGCCG |
| M8-F | CGAGCTCTGATTAATTAATTCAGAACGCCCGGCTTCGGCCG |
| M9-F | CGAGCTCTGATTAATTAATTCAGAACGCCCGGCUAACGCCGGGCGTTTTTTATGCA |
| M10-F | CGAGCTCTGATTAATTAATTCAGAACGCCCGGCAAATCGCCGGGCGTTTTTTATGCA |
| M11-F | CGAGCTCTGATTAATTAATTCAGAACGCCCGGCAAAAGCCGGGCGTTTTTTATGCA |
| RNase-F | AAGGAAAAAAGCGGCCGCGAATTAATTAATCAAAGGAGATCAATACAAATAAT |
| RNase-R | AAGGAAAAAAGCGGCCGCATTATTTGTATTGATCTCCTTTACTATCTCTCGA |
表选项
1.2 Construction of characterization devicesThe development of devices that allow the characterization of our new BioBrick terminator variants was based on construction of the GFP/RFP dual fluorescent system being with the inputs/outputs to the system controlled by the P43 promoter. We started by introducing several additional restriction enzyme recognition sites into the multiple cloning site (MCS) of the pP43NMK expression vector. Firstly, a gfp reporter gene fragment was amplified using primers GFP-F/GFP-R (Table 2). The resulting PCR product was digested with KpnⅠ and PstⅠ and ligated intramolecularly to generate plasmid pP43NMK-gfp, which contained successive SacⅠ, XbaⅠ, SpeⅠ and PstⅠ recognition sites downstream gfp. Secondly, a fragment of the rfp reporter gene was amplified from the pUC57-simple plasmid using primers RFP-F/RFP-R and digested with SacⅠ and SpeⅠ. The resulting PCR product was ligated in pP43NMK-gfp plasmid digested with the same enzymes (SacⅠ and SpeⅠ) to generate a control device pP43NMK-gfp-rfp. The next step was the insertion of a testing terminator genetic sequence which was synthesized and amplified using primers λto-F/λto-R to generate pP43NMK-gfp-λto-rfp. The construction of an insulated terminator was performed by insertion of new genetic devices (RNase site and strong hairpin) in the testing plasmid by means of amplification of pUC57-simple using primers RNase F/RNase R. The resulting PCR product was immediately mixed with 1 U of DpnⅠ to digest the remaining circular plasmid and the product was digested with NotⅠ then self-ligated to generate the plasmid pUC57-RNase site-λto-RNase site-strong hairpin. From the resulting plasmid, we amplified the fragment harboring the RNase site, λtoterminator and strong hairpin with primers λto-F/λto-R. The resulting PCR fragment was digested with SacⅠ and SpeⅠ, then ligated downstream of the gfp reporter in pP43NMK-gfp digested with the same enzymes to generate the testing device pP43NMK-gfp-RNase site-λto-RNase site-strong hairpin-rfp. Furthermore, we attained our characterization construct model by constructing terminator variants M1, M2, M3, M4, M5, M6, M7, M8, M9, M10 and M11 using primers M1-F/λto-R, M2–F/λto-R, M3-F/λto-R, M4-F/λto-R, M5-F/λto-R, M6-F/λto-R, M7-F/λto-R, M8-F/λto-R, M9-F/λto-R, M10-F/λto-R and M11-F/λto-R to generate recombinant plasmids named pP43NMK-gfp-M1-rfp, pP43NMK-gfp-M2-rfp, pP43NMK-gfp-M3-rfp, pP43NMK-gfp-M4-rfp, pP43NMK-gfp-M5-rfp, pP43NMK-gfp-M6-rfp, pP43NMK-gfp-M7-rfp, pP43NMK-gfp-M8-rfp, pP43NMK-gfp-M9-rfp, pP43NMK-gfp-M10-rfp and pP43NMK-gfp-M11-rfp, respectively. Recombinant bacteria harboring controls and testing devices were isolated, and the entire resulting devices were confirmed by sequencing. We further continued by transforming all confirmed plasmids into B. subtilis 168 competent cells, and finally we obtained a control strain B. subtilis 168 GFP-RFP and recombinant test strains B. subtilis 168-M1, B. subtilis 168-M2, B. subtilis 168-M3, B. subtilis 168-M4, B. subtilis 168-M5, B. subtilis 168-M6, B. subtilis 168-M7, B. subtilis 168-M8, B. subtilis 168-M9, B. subtilis 168-M10, B. subtilis 168-M11 and B. subtilis 168 GFP-RNase site-λto-RNase site-strong hairpin-rfp which were treated and subjected to the characterization experiment.
1.3 Media and culture conditionsE. coli JM109, TOP10 and B. subtilis 168 were typically cultured in LB broth (per liter: 10.0 g tryptone, 5.0 g yeast extract, and 10.0 g NaCl) at 37 ℃ with shaking speed of 220 r/min. Solid media were prepared by adding 1.5 g/Lagar to the respective media. Unless otherwise stated, the antibiotic concentrations were 100 mg/Lampicillin for E. coli, and 100 mg/Lkanamycin for B. subtilis 168. To evaluate the expression of our genetic devices, all plasmids harboring genetic sequences were cultured in LB medium supplemented with antibiotic and flask cultivations were performed at 37 ℃ with a rotational speed of 220 r/min for 12 hours where samples were collected and treated every 2 hours and subjected to fluorescence measurements.
1.4 Analytical methods1.4.1 GFP and RFP intensity quantificationFor the GFP and RFP determination, the culture broth was centrifuged at 10 000×g for 10 min, washed with 0.1 mol/L phosphate-buffered saline (pH 7.4), and diluted in the same buffer. Therefore, 200 μL of each sample was pipetted, added to the 96-well plates and immediately subjected to fluorescence determination. The GFP and RFP fluorescence intensity produced by the characterization devices were measured by using a Cytation 3 Cell Imaging Multi-Mode Reader (BioTek) at an excitation/emission wavelength of 490/530 nm and 555/584 nm respectively.
1.4.2 Analysis of GFP mRNA transcript levelSamples taken from the culture media between 7 and 8 hours were thawed and centrifuged for 2 min at~11 000×g at 4 ℃. The supernatant was discarded, and the pellet was immediately frozen at ?80 ℃. Cell pellets were suspended in Tris-EDTA (TE) buffer (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA) with lysozyme to a final concentration of 0.4 mg/ml. Total RNA was extracted with RNAprep Pure Kit (Tiangen biotech, CHINA) and RNA concentration was determined with the NanoDrop1000 (Thermo Fisher Scientific, MA, USA). Furthermore, cDNA was synthesized by Takara Prime Script Reverse Transcriptase with gDNA eraser and quantitative-PCR (Q-PCR) was carried out using TaKaRa SYBR Premix Dimer Eraser.
2 Results and discussion2.1 Structures of the λto terminator and its variantsThe DNA sequences as well as the free energies of the λtoterminator [28] and its variants are shown in Table 3. In particular, both the RNA hairpin structures and their corresponding free energies were predicted by KineFold [29]. The loop and the adjacent C-G base pair were examined and mutated since each contributed to the terminator strength[9]. Generally, nucleotides mismatches occur very rarely in strong terminators, thus, the nucleotides mismatches in the stem of the hairpin were eliminated. In addition, the effects of the poly (T) tail was also analyzed. Specifically, the TGCC loop in variants M2, M3, M4, M5, M6, M7 and M8 were replaced with TTCG and GAAA which are considered to increase terminator efficiency and mRNA stability in many prokaryotes [7]. In variants M1, M2, M3 and M4, A was substituted with T to increase the number of T in the poly (T) tract but in variants M9, M10 and M11, the TGCC loop was substituted with GAAC, AAATC and AAAA, respectively. Additionally, to eliminate the mismatched nucleotide pair, T at positions 22 and 26 in the G+C rich stem were substituted with C in the variants M6, M7, M8, M9, M10 and M11 (Fig. 1).
Table 3 Wild type and terminator variant sequences used in this study
| Names | Wild-type and terminators variants DNA sequences | Free energy (kcal/mol) |
| WT λto | TGATTAATTAATTCAGAACGCTCGGTTGCCGCCGGGCGTTTTTTATGCA | ?17.5 |
| M1 | TGATTAATTAATTCAGAACGCTCGGTTGCCGCCGGGCGTTTTTTTTGCA | ?17.5 |
| M2 | TGATTAATTAATTCAGAACGCTCGGTTTCGGCCGGGCGTTTTTTTTGCA | ?19.3 |
| M3 | TGATTAATTAATTCAGAACGCTCGGTTTCGGCCGGGCGTTTTTTTTGCA | ?19.3 |
| M4 | TGATTAATTAATTCAGAACGCTCGGTGAAAGCCGGGCGTTTTTTTTGCA | ?20.4 |
| M5 | TGATTAATTAATTCAGAACGCTCGGTGAAAGCCGGGCGTTTTTTATGCA | ?20.4 |
| M6 | TGATTAATTAATTCAAAACGCCCGGCGAAAGCCGGGCGTTTTTTATGCA | ?21.7 |
| M7 | TGATTAATTAATTCAGAACGCCCGGCGAAAGCCGGGCGTTTTTTATGCA | ?24.3 |
| M8 | TGATTAATTAATTCAGAACGCCCGGCTTCGGCCGGGCGTTTTTTATGCA | ?23.5 |
| M9 | TGATTAATTAATTCAGAACGCCCGGCGAACGCCGGGCGTTTTTTATGCA | ?22.0 |
| M10 | TGATTAATTAATTCAGAACGCCCGGCAAATCGCCGGGCGTTTTTTATGCA | ?22.0 |
| M11 | TGATTAATTAATTCAGAACGCCCGGCAAAAGCCGGGCGTTTTTTATGCA | ?21.5 |
表选项
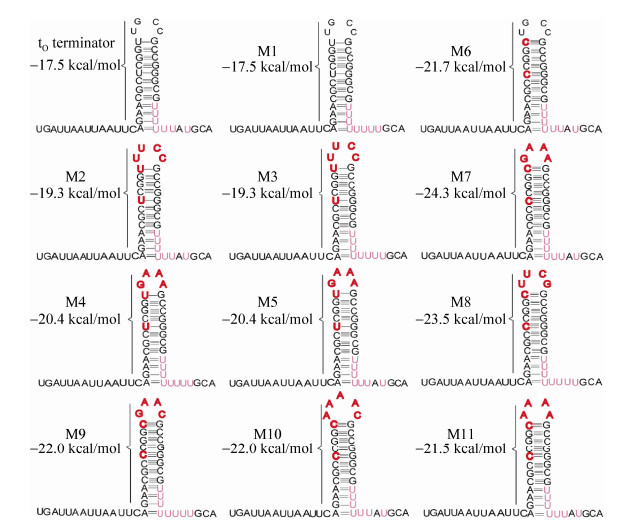 |
| Figure 1 Secondary structures of the λtO terminator and its variants. The predicted stabilities of the RNA hairpin structures were determined by the Kinefold web server. The termination positions for all engineered functional λtO variants were highlighted in red. |
| 图选项 |
2.2 Termination efficiencies of the constructed terminator variantsTo assess the effects of the loops, poly (T) tract and mismatched nucleotide pair on transcription termination, as well as the termination efficiencies of the variants were determined by measuring the input and output fluorescence of the reporter genes (gfp and rfp) in the constructed system (Fig. 2). As shown in Fig. 3B, the insertion of the wild-type strong λtoterminator and its variants resulted in low expression of rfp, suggesting the strong termination efficiency of all the terminators. After further quantification of the GFP fluorescence (Fig. 3A) and calculation [18], terminator variants M3, M4 and M11 showed higher termination efficiency values compared with the wild type λto and the remaining variants (Fig. 3C). Interestingly, the inclusion of RNase sites in the variant M12 (Fig. 3C), showed a considerable termination efficiency of about 98% which prove its positive effect on the termination efficiency. The results clearly showed that the loop causes no significant effect on transcription termination although both tetra loops GAAA and TTCG are frequently found in many prokaryotic and eukaryotic RNAs [2-7]. Meanwhile, the results also suggested that the introduction of mutations to enhance the RNA stem stability did not contribute to the termination efficiency, which are different from that in E. coli[22].
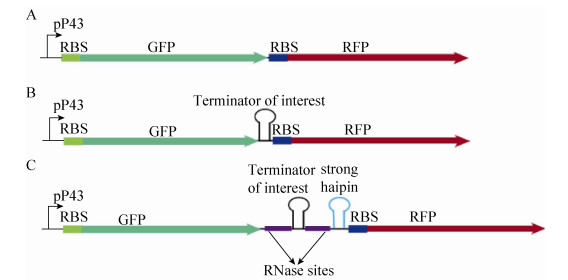 |
| Figure 2 Structures of the expression cassettes with two reporter genes gfp and rfp. (A) The control without terminator. (B) Terminator was inserted between two fluorescent protein encoding genes. (C) RNase sites and strong hairpin were inserted followed by rfp gene. RBS: ribosome-binding site. |
| 图选项 |
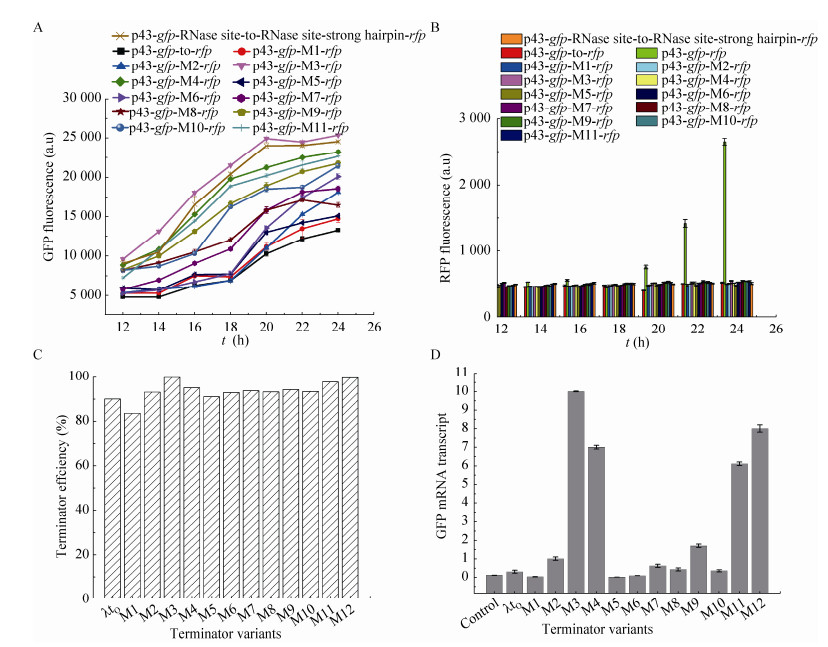 |
| Figure 3 Comparison of termination efficiency and mRNA stability of the λto terminator variant. (A) The GFP fluorescence of the native terminator (control) and the variants. (B) The RFP fluorescence of the native terminator and its variants. (C) Termination efficiencies of native (λtO) and its variants. (D) The mRNA levels of the gfp gene with the λtO terminator and its variants. Error bars are standard deviations calculated from three independent experiments. |
| 图选项 |
2.3 Insertion of terminator increased upstream gene expressionIn addition to evaluating the termination efficiency, the effect of the terminators on the upstream gene was also investigated and compared. Furthermore, the low gfp expression revel obtained by measuring the negative control compared with other studied terminator variants (Fig. 3A), explains the role of terminators on the upstream gene. Interestingly, the GFP fluorescence was significantly increased when inserting all the terminator variants (Fig. 3A), indicating its positive effect on the RNA stability of the upstream gfp, which is similar to previous studies in E. coli [10, 30] and S. cerevisiae[9, 13]. To further confirm this phenomenon, the mRNA of gfp in all the constructs were analyzed. Compared with the control, most of the constructs with terminator variants (especially M2, M3, M4, M9, M11 and M12) yielded a higher level of gfp mRNA (Fig. 3D). Recently, it has been reported that the addition of RNase sites surrounding the target terminator can impact the expression of reporter genes in E. coli [8, 31]. Thus, two RNase sites and one strong hairpin were inserted between the reporter genes gfp and rfp (Fig. 2C). As expected, the expression of gfp was significantly increased (Fig. 3A).
3 ConclusionsIn this study, the hairpin and the U-rich sequence of the strong bacteriophage λto terminator were first engineered and investigated in Bacillus subtilis 168. Compared with the wild-type strong λto terminator, all the variants showed a higher termination efficiencies. The results suggested that the hairpin loop is not a critical factor for strong terminator. More importantly, the results first demonstrated that terminator insertion can significantly increase the expression of upstream gene in B. subtilis. As a result, these rho-independent terminators should be used as a potential tool for synthetic biology research in B. subtilis. In addition, the constructed terminators should also be used for increasing mRNA stability and the corresponding enzyme production in B. subtilis.
参考文献
| [1] | Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake.Genome Biol,2007, 8(2): R22.DOI: 10.1186/gb-2007-8-2-r22 |
| [2] | Lesnik EA, Sampath R, Levene HB, et al. Prediction of rho-independent transcriptional terminators in Escherichia coli.Nucleic Acids Res,2001, 29(17): 3583–3594.DOI: 10.1093/nar/29.17.3583 |
| [3] | Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3'-end chronicles.J Mol Biol,2011, 412(5): 793–813.DOI: 10.1016/j.jmb.2011.03.036 |
| [4] | Abe H, Aiba H. Differential contributions of two elements of rho-independent terminator to transcription termination and mRNA stabilization.Biochimie,1996, 78(11-12): 1035–1042.DOI: 10.1016/S0300-9084(97)86727-2 |
| [5] | Abe H, Abo T, Aiba H. Regulation of intrinsic terminator by translation in Escherichia coli: transcription termination at a distance downstream.Genes Cells,1999, 4(2): 87–97.DOI: 10.1046/j.1365-2443.1999.00246.x |
| [6] | Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions.Bioessays,2002, 24(8): 700–707.DOI: 10.1002/(ISSN)1521-1878 |
| [7] | d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators: a statistical analysis of their RNA stem-loop structures.J Mol Biol,1990, 216(4): 835–858.DOI: 10.1016/S0022-2836(99)80005-9 |
| [8] | Cambray G, Guimaraes JC, Mutalik VK, et al. Measurement and modeling of intrinsic transcription terminators.Nucleic Acids Res,2013, 41(9): 5139–5148.DOI: 10.1093/nar/gkt163 |
| [9] | Curran KA, Morse NJ, Markham KA, et al. Short synthetic terminators for improved heterologous gene expression in yeast.ACS Synth Biol,2015, 4(7): 824–832.DOI: 10.1021/sb5003357 |
| [10] | Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin.Proc Natl Acad Sci USA,1995, 92(19): 8793–8797.DOI: 10.1073/pnas.92.19.8793 |
| [11] | Li R, Zhang Q, Li JB, et al. Effects of cooperation between translating ribosome and RNA polymerase on termination efficiency of the Rho-independent terminator.Nucleic Acids Res,2016, 44(6): 2554–2563.DOI: 10.1093/nar/gkv1285 |
| [12] | Britton RA, Lupski JR. Functional analysis of mutations in the transcription terminator T1 that suppress two dnaG alleles in Escherichia coli.Mol Gen Genet,1995, 246(6): 729–733.DOI: 10.1007/BF00290719 |
| [13] | Curran KA, Karim AS, Gupta A, et al. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications.Metab Eng,2013, 19: 88–97.DOI: 10.1016/j.ymben.2013.07.001 |
| [14] | Chen YJ, Liu P, Nielsen AAK, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints.Nat Methods,2013, 10(7): 659–664.DOI: 10.1038/nmeth.2515 |
| [15] | Bonnet J, Yin P, Ortiz ME, et al. Amplifying genetic logic gates.Science,2013, 340(6132): 599–603.DOI: 10.1126/science.1232758 |
| [16] | Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells.Nat Biotechnol,2013, 31(5): 448–452.DOI: 10.1038/nbt.2510 |
| [17] | Gasanov NB, Toshchakov SV, Georgiev PG, et al. The use of transcription terminators to generate transgenic lines of chinese hamster ovary cells (CHO) with stable and high level of reporter gene expression.Acta Naturae,2015, 7(3): 74–80. |
| [18] | Lin MT, Wang CY, Xie HJ, et al. Novel utilization of terminators in the design of biologically adjustable synthetic filters.ACS Synth Biol,2016, 5(5): 365–374.DOI: 10.1021/acssynbio.5b00174 |
| [19] | Pfleger BF, Pitera DJ, Smolke CD, et al. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes.Nat Biotechnol,2006, 24(8): 1027–1032.DOI: 10.1038/nbt1226 |
| [20] | Ito Y, Yamanishi M, Ikeuchi A, et al. Characterization of five terminator regions that increase the protein yield of a transgene in Saccharomyces cerevisiae.J Biotechnol,2013, 168(4): 486–492.DOI: 10.1016/j.jbiotec.2013.09.024 |
| [21] | Fritsch TE, Siqueira FM, Schrank IS. Intrinsic terminators in Mycoplasma hyopneumoniae transcription.BMC Genomics,2015, 16(1): 273.DOI: 10.1186/s12864-015-1468-6 |
| [22] | de Hoon MJL, Makita Y, Nakai K, et al. Prediction of transcriptional terminators in Bacillus subtilis and related species.PLoS Comput Biol,2005, 1(3): e25.DOI: 10.1371/journal.pcbi.0010025 |
| [23] | Hess GF, Graham RS. Efficiency of transcriptional terminators in Bacillus subtilis.Gene,1990, 95(1): 137–141.DOI: 10.1016/0378-1119(90)90425-Q |
| [24] | Harwood CR, Cranenburgh R. Bacillus protein secretion: an unfolding story.Trends Microbiol,2008, 16(2): 73–79.DOI: 10.1016/j.tim.2007.12.001 |
| [25] | Liu YF, Liu L, Shin HD, et al. Pathway engineering of Bacillus subtilis for microbial production of N-acetylglucosamine.Metab Eng,2013, 19: 107–115.DOI: 10.1016/j.ymben.2013.07.002 |
| [26] | Tsukahara K, Ogura M. Characterization of DegU-dependent expression of bpr in Bacillus subtilis.FEMS Microbiol Lett,2008, 280(1): 8–13.DOI: 10.1111/fml.2008.280.issue-1 |
| [27] | Zhang JJ, Kang Z, Ling ZM, et al. High-level extracellular production of alkaline polygalacturonate lyase in Bacillus subtilis with optimized regulatory elements.Bioresour Technol,2013, 146: 543–548.DOI: 10.1016/j.biortech.2013.07.129 |
| [28] | Zhang XZ, Yan X, Cui ZL, et al. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis.Nucleic Acids Res,2006, 34: e71.DOI: 10.1093/nar/gkl358 |
