王玉芳1, 蔡元锋2, 侯扶江1, 贾仲君2, Bowatte Saman1


1. 兰州大学草地农业生态系统国家重点实验室, 兰州大学农业农村部草牧业创新重点实验室, 兰州大学草地农业科技学院, 甘肃 兰州 730020;
2. 中国科学院南京土壤研究所, 土壤与农业可持续发展国家重点实验室, 江苏 南京 210008
收稿日期:2018-11-03;修回日期:2019-01-07;网络出版日期:2019-04-22
基金项目:国家自然科学基金(31672472,41401294);****和创新团队发展计划(IRT-17R250);甘肃省退牧还草科技支撑(GH2017-TMHC001)
作者简介:Saman Bowatte is a Professor at college of Pastoral Agriculture Science and Technology (CPAST) of Lanzhou University, China. Professor Bowatte received PhD in Soil Science from the Massey University, New Zealand in 2003, then moved to Nagoya University, Japan for Post-Doctoral fellowship before returned to Grasslands campus of AgResearch New Zealand in 2006 to work as a Soil Microbial Ecologist for 10 years. In March 2016 he joined Lanzhou University. The main theme of his research has been focused on plant-soil-microbe interactions towards on better understanding and manipulating of the grazed pastures to improve grasslands productivity while minimising environmental concerns. It includes (a) nitrogen dynamics in pastoral soils (b) climate change impacts on grasslands (c) greenhouse gas emissions and mitigation (d) molecular understanding of soil microbial diversity and function. Professor Bowatte has made significant contribution towards the microbial mechanisms underlying the ecological consequence of plant-soil-microbe interactions on nitrogen cycling and nitrous oxide production from livestock farming. His research demonstrated not only plant identity influence N2O production in grassland soils but also grassland plants itself emit N2O and showed that bacteria on the grass leaf surfaces can be a previously unrecognised source of N2O from grasslands. Most recently, his research has established strong links between the community assembly of N2O-emitting bacteria and grassland plant species, and identified the remarkable possibility through the grass seed microbiome to reduce N2O emission. He has > 40 peer-reviewed publication including Nature Geoscience, ISME Journal, Soil Biology and Biochemistry, Agriculture Ecosystem and Environment and Geoderma.
*通信作者:Saman Bowatte, Tel:+86-931-8911955, E-mail:samanbowatte@lzu.edu.cn.
摘要:[目的] 针对我国甘肃三个典型生态区草地土壤(玛曲MQ、临泽LZ和环县HX),研究其甲烷氧化潜力、甲烷氧化菌(methane-oxidizing bacteria,MOB)丰度及可能存在的群落分异规律。[方法] 通过原位分析、室内高浓度甲烷模拟培养三种典型土壤及实时荧光定量、高通量测序的方法研究甲烷氧化菌标靶基因pmoA序列的组成及其丰度变化规律。[结果] 三种典型草地土壤的原位甲烷氧化菌的丰度存在显著差异,表现为MQ > HX > LZ,其数量范围为为0.18-6.86×107g/d.w.s.;甲烷氧化潜力也表现出类似规律,其通量为109-169 mg/(m2·h);甲烷氧化潜力与原位土壤中甲烷氧化菌丰度有正相关。三种草地土壤甲烷氧化菌存在明显的空间异质性,采用高通量测序的方法,发现三种草地原位土壤中的优势类群为USCγ(Upland Soil Cluster gamma,USCγ);然而,室内高浓度甲烷氧化过程中,传统的甲烷氧化菌均发生明显增加,MQ土壤中TypeⅡ的Methylocystis为优势类群,而LZ和HX土壤的优势类群均为TypeⅠ型Methylosarcina。[结论] 这些研究结果表明,我国甘肃典型草地土壤中也存在难培养的大气甲烷氧化菌和经典的可培养甲烷氧化菌,这些微生物极可能氧化极低浓度的大气甲烷,也可能利用闭蓄于土壤中的高浓度甲烷生长。未来应采用先进技术原位观测大气甲烷氧化过程并分离相应微生物类群,研究草地土壤甲烷氧化菌地理分异规律及其环境驱动机制。
关键词:草地甲烷氧化pmoA基因
Aerobic methane oxidation in typical grassland soil
Yufang Wang1, Yuanfeng Cai2, Fujiang Hou1, Zhongjun Jia2, Saman Bowatte1


1. State Key Laboratory of Grassland Agro-ecosystems; Key Laboratory of Grassland Livestock Industry Innovation, Ministry of Agriculture and Rural Affairs; College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou 730020, Gansu Province, China;
2. State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing 210008, Jiangsu Province, China
*Corresponding author: Saman Bowatte, Tel:+86-931-8911955, E-mail:samanbowatte@lzu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31672472, 41401294), by the Changjiang Scholars and Innovation Team Development Program (IRT-17R250) and by the Gansu Province's Grazing and Grazing Technology Support (GH2017-TMHC001)
Abstract: [Objective] The soil methane uptake potential, abundance and community structure of methane-oxidizing bacteria were investigated in three grasslands located in three different ecoregions:Maqu, Linze and Huanxian of Gansu province of China. [Methods] Eight days incubation of soil with elevated concentration of methane was carried out to measure methane uptake capacity. The methane-oxidizing bacteria abundance was quantified by real time quantitive PCR targeting particulate methane monooxygenase coding gene (pmoA) in soils. The methane-oxidizing bacteria community structure was assessed by amplicon MiSeq sequencing. [Results] The potential of methane oxidation of three grassland soils ranged from 108.53±13.12 to 168.87±18.57 mg/(m2·h). The abundance of methane-oxidizing bacteria ranged from 1.76×107 to 6.86×107 pmoA gene copies g/d.w.s. The methane oxidation potential was positively correlated with methane-oxidizing bacteria abundance at day 0 (R2 = 0.5537). MiSeq sequencing analysis revealed significant spatial heterogeneity of methane-oxidizing bacteria community within the same grassland type. The Upland Soil Cluster gamma belonging to uncultured atmospheric methane oxidizers was the dominant methanotrophic lineage within methane-oxidizing bacteria gene types found in situ grassland soils. However, the conventional methane-oxidizing bacteria increased significantly after incubated these soils under high concentration methane, such as Methylocystis in Maqu soil and Methylosarcina in Linze and Huanxian soils. [Conclusion] Both uncultured atmospheric methane-oxidizing bacteria and the conventional methane-oxidizing bacteria may play an important role in the process of methane oxidation in the typical grassland soils in Gansu province of China. These microbes are very likely to oxidize the trace methane in atmosphere, and may also grow with high concentration of methane that stored in the soil. In the future, advanced techniques should be used to observe the atmospheric methane oxidation process in situ and to isolate the corresponding microbial groups, and finally reveal the geographical differentiation of methane-oxidizing bacteria in grassland soils and the environmental driving mechanism.
Keywords: grasslandmethane oxidationpmoA gene
甲烷(CH4)是一种重要的温室气体,对温室效应的贡献为17%,仅次于CO2。目前,大气中的CH4平均浓度达到1.867 μL/L (part per million by volume)[1],比工业革命前大气甲烷平均浓度0.72 μL/L[2]高出259%。据IPCC估算,全球土壤可氧化大气中甲烷约为30 Tg/Yr[3]。因此,土壤作为大气CH4汇是全球气候变化研究的热点之一。
森林和草地土壤是重要的大气CH4汇[4-5],其中难培养甲烷氧化菌被认为是在其中发挥了主要作用。我国草原占国土面积的40%,占世界草地总面积的16%。据估算,中国草地土壤平均消耗甲烷量为0.168–1.132 Tg CH4/Yr (平均0.65 Tg CH4/Yr),平均速率为59.62 mg CH4/(m2·h)。同时,草地甲烷氧化存在明显的地理分异规律,如青藏高原高寒地区、东北干旱半干旱地区和东部湿润半湿润地区的草地甲烷吸收量分别为0.284 Tg CH4/Yr、0.204 Tg CH4/Yr和0.162 Tg CH4/Yr[6]。长期以来,由于技术手段的限制,影响我国草地土壤甲烷氧化通量研究主要侧重于环境要素的耦合分析,如土壤湿度[6]、温度[7]、氮添加[8-11]、放牧强度[12]等,鲜有甲烷氧化菌的微生物机制报道。
近年来随着高通量测序等先进技术的快速发展,草地土壤甲烷氧化菌生理生态学研究得到了国内外****的高度关注。甲烷氧化菌广泛分布于地球生态系统,根据其代谢途径、形态结构和生理特征等差异,可将已知的好氧甲烷氧化菌主要分为TypeⅠ、TypeⅡ两类。TypeⅠ型主要属于γ-proteobacteria纲,TypeⅡ型主要属于α-proteobacteria纲。另外,最近发现存在极端嗜酸的好氧甲烷氧化菌Verrucomicrobia门[13]。然而,目前发现的所有可培养好氧甲烷氧化菌底物亲和力较低,这类微生物通常栖息于甲烷浓度较高的环境(几千到上万μL/L),如湖泊和稻田湿地,仅能通过氧化高浓度甲烷繁殖生长。大气中甲烷浓度仅为1.85 μL/L左右,因此土壤大气甲烷氧化菌需要具备极高的底物亲和力。理论计算表明,已知的所有可培养甲烷氧化菌均无法利用如此低浓度甲烷生长,分子生态学研究表明,目前已知的所有能够氧化大气甲烷的土壤中均存在大气甲烷氧化菌,如草地[14]、森林[15]、沙漠[16]等土壤,但目前尚未有大气甲烷氧化菌的纯培养物。针对已知的甲烷氧化菌,Holmes等[17]研究了其单加氧酶功能基因pmoA的系统发育进化关系,并设计了特异性引物;进一步通过构建pmoA基因克隆文库的方法,研究了森林土壤中甲烷氧化菌,发现了特异的pmoA基因序列并将其定义为RA14和RA21,推测这些微生物不同于传统的甲烷氧化菌[18];随后,Knief等研究了土壤大气甲烷氧化潜力,将这些难培养大气甲烷氧化菌命名为USCα (upland soil cluster α)和USCγ (upland soil cluster γ)[19]。Ma等采用pmoA基因克隆技术,研究发现我国内蒙典型草原土壤中USCγ是大气甲烷氧化菌的优势类群[20]。Kou等针对我国21个不同草地开展了甲烷氧化菌的生物地理学研究,发现土壤中同时栖息着大量的传统甲烷氧化菌[21]。然而,这些传统甲烷氧化菌能否利用高浓度甲烷,以及是否与难培养大气甲烷共存,目前的相关研究报道较少。据此,本研究针对我国甘肃3个不同生态区的典型草地采集土壤样本,通过室内高浓度甲烷培养和高通量测序的手段对其CH4氧化潜力、甲烷氧化菌丰度和多样性进行分析研究,揭示不同类型草地土壤甲烷氧化潜力、甲烷氧化菌丰度及群落组成规律,以期为草地生态系统甲烷汇的评估提供基础数据支撑。
1 材料和方法 1.1 土壤样本采集 土壤样品在2016年9月23日至10月5日期间采集于中国甘肃省3个不同生态区的草地。每个采样点的土壤特征描述列于表 1中。在每个草地类型中随机选择3个10×10 m的小区,每个小区之间至少相隔50 m。在每个取样小区内用直径10 cm的土钻随机取10个0–10 cm深度土柱,混合后作为一个土壤样品。因此,每个草地类型包含3个重复土壤样本,用冷藏盒将土壤样本带回实验室。在24 h内,用2 mm土筛去除土壤样品中的植物残体,石头等。将样品分为2份,一份储存在–20 ℃用于DNA提取,一份风干后储存在4 ℃冷库,用于土壤理化性质测定和后续培养实验。MQ、LZ和HX的土壤理化性质如下:土壤水分含量分别为(17.41±0.90)%、(11.71±0.75)%和(5.41±0.40)%;pH分别为6.33±0.15、7.84±0.13和8.59±0.12;全碳分别为65.2±9.1 g/kg、27.2±9.2 g/kg和14.2±1.3 g/kg;全氮分别为5.5±0.7 g/kg、1.4±0.3 g/kg和0.6±0.1 g/kg。原位土壤样本保存后,用于随后土壤甲烷氧化菌数量和组成分析,在随后实验中作为Day 0零时刻样品。
表 1. 采样地点基本概况描述 Table 1. Basic information of sampling site
| Grassland identity | MQ | LZ | HX |
| Ecoregion | Alpine meadow Qinghai-Tibet Plateau | Northwest inland arid region | Semi-arid Loess Plateau |
| Sampling location | Maqu county 35° 58’ N, 101° 53’ E | Linze county 39° 11’ N 100° 6’ E | Huanxian county 37° 12’N, 106° 82’ E |
| Soil type | Subalpine meadow soil | Salinized meadow soil | Yellow soil |
| Average annual rainfall | 620 mm | 112.9 mm | 359.3 mm |
| Altitude | 3650 m | 1400 m | 1650 m |
| Annual average temperature | 1.2 ℃ | 7.6 ℃ | 7.1 ℃ |
| Dominant plant species | Carex kansuensis/Festuca ovina Poa poophagorum/Elymus nutans | (Sowing pasture) Medicago sativa Linn. Festuca elata | Stipabungeana/Artemisia capillari Lespedeza davurica |
表选项
1.2 室内模拟甲烷氧化微生物过程 根据郑燕等[22]描述的方法进行。简要说明如下:将新鲜(相当于6 g风干土壤)土壤置于120 mL血清瓶中,每种土壤类型有3个生物重复。调节土壤的含水量达到最大持水量的40%,在28 ℃避光预培养5 d,使土壤中的微生物恢复活性。然后,再次调整土壤含水量至最大持水量的60%,用黑色丁基橡胶塞密封并用铝盖封口。从每个密封瓶子中用注射器取出1.2 mL空气,然后注入相同体积的CH4 (99.9%)。这使得每个瓶子的顶部空间在培养开始时含有1% (10000 μL/L)的CH4浓度,然后在28 ℃避光培养。在培养开始后的第1、3、5和8天,从每个瓶中取出0.25 mL空气,用氩气稀释100倍并通过气相色谱(GC-7890A,America)检测CH4浓度的动态变化,8 d后结束培养,收集土壤样品并储存于–20 ℃以进行DNA提取。草地土壤甲烷氧化潜力的计算,主要根据培养瓶内甲烷浓度降低最快的时间点推算,并将之换算为标准状态。
1.3 土壤DNA提取 采用FastDNA Spin Kit for Soil (MP Bio)试剂盒按照说明书完成操作。统一提取原位土壤样品和室内模拟培养结束后的样品,获得土壤样品总DNA后,将其提取的DNA溶解于100 μL的DNA洗脱液中。通过微量紫外分光光度计(NanoDrop ND-1000)测定DNA浓度和纯度,同时利用1.2%的琼脂糖凝胶电泳分析DNA的完整性。
1.4 实时荧光定量PCR测定甲烷氧化菌的丰度 使用CFX96 Real-Time PCR System (Bio-Rad公司)对土壤总DNA中的甲烷氧化菌功能pmoA基因进行定量,qPCR引物和扩增条件如表 2所示。获得上述各微生物标靶基因的重组质粒,然后10倍梯度稀释用于标准曲线绘制。定量PCR反应体系如下:10 μL SYBR Premix Ex Taq (TaKaRa),0.35 μL引物(10 μmol/L),1.0 μL DNA模板,加入DNase/RNase-free水补足至20 μL。在所有分析中,灭菌的超纯水代替土壤DNA作为空白对照。定量PCR的扩增效率在90%左右,标准曲线R2为0.996。
表 2. 本研究中用到的引物信息及反应条件 Table 2. Primers and PCR thermal cycle conditions used in this study
| Primer name | Primer sequence (5’→3’) | Thermal profile | Molecular analysis | Reference |
| A189F mb661R | GGNGACTGGGACTTCTGG CCGGMGCAACGTCYTTACC | 95 ℃, 3.0 min; 40×(95 ℃, 10 s; 55 ℃, 30 s; 72 ℃, 30 s; 80 ℃, 5 s; with plate read); melt curve 65 ℃ to 95 ℃, incremental 0.5 ℃, 0.5 s+plate read | qPCR | [14] [17] |
| A189F A682R | GGNGACTGGGACTTCTGG GAASGCNGAGAAGAASGC | 95 ℃, 3.0min; 32×(95 ℃, 10 s; 55 ℃, 30 s; 72 ℃, 30 s); 72 ℃, 10 min | MiSeq Sequencing | [23] |
表选项
1.5 Illumina Miseq测序 通过Illumina Miseq测序平台分析甲烷氧化菌的相对丰度和微生物组成。利用引物A189F/A682R (表 2)扩增pmoA基因,并将6 bp样品特异性接头序列连接至A189F引物的5?末端。50 μL PCR混合体系包括:25 μL TaKaRa Premix Ex Taq,1.0 μL引物(10 μmol/L),2 μL DNA模板,加入DNase/ RNase-free水补足至50 μL反应体系。获得扩增产物后,利用Agarose Gel DNA Fragment Recovery Kit Ver.2.0试剂盒(TaKaRa)将PCR产物切胶纯化,纯化产物溶解于30 μL DNase/RNase-free H2O,将3.0 μL的PCR产物通过1.2%琼脂糖凝胶电泳检测纯化效果,利用微量紫外分光光度计(NanoDrop? ND-1000 UV-Vis)测定扩增产物的浓度。将不同样品的PCR纯化产物等摩尔数混合后,使用TruSeq Nano DNA LT样品制备试剂盒组A (24个样品)构建测序文库,并且使用MiSeq试剂盒v3 (600个循环)进行测序。使用Mothur软件(版本1.33.3)对MiSeq测序结果读取、合并和质量过滤。使用'make.contigs'命令(deltaq = 5)将双端序列合并,并且'trim.seqs'命令将引物标签去除和长度筛选(留下长度在420–500 bp之间的序列)。最后,使用'classify.seqs'命令将高质量序列直接分类[24] (总共114912)。pmoA高通量测序原始数据已经提交到NCBI的SRA数据库,登录号SRP154541。
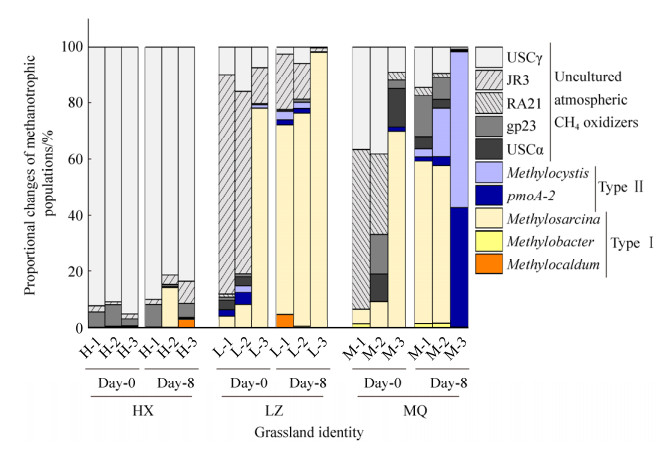
1.6 数据分析 CH4氧化量的计算参考文献[25],用公式(1)计算:
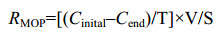 | 公式(1) |
进一步根据培养瓶内甲烷氧化量,结合培养瓶内土壤厚度0.5 cm,假设原位0–5 cm厚度的土壤具有相似的甲烷吸收速率,以此估算原位5 cm土层的甲烷氧化通量。
所有数据采用SPSS 20.0进行处理分析,处理之间的平均值差异采用one-way ANOVA单因素方差分析,P < 0.05表示显著差异,使用OriginPro 2016进行作图。
2 结果和分析 三种草地土壤中,原位甲烷氧化菌的基因拷贝丰度表现出明显的差异;MQ、HX和LZ草地土壤中pmoA基因拷贝数分别为4.46×107 g/d.w.s.、2.30×107 g/d.w.s.、7.63×106 g/d.w.s.,表现为MQ > HX > LZ。高浓度甲烷室内培养条件下,土壤甲烷氧化潜力也得到类似结果(图 1-A),发现第1天时氧化最快,随后逐渐放缓。以第1天的甲烷氧化速率表征土壤的甲烷氧化最大潜力,发现MQ土壤的氧化潜力最大,约为(168.87±18.57) mg/(m2·h);HX次之,约为(124.68±21.87) mg/(m2·h);LZ最低,为(108.53±13.12) mg/(m2·h),三种土壤的甲烷氧化潜力之间存在显著性的差异(图 1-B);室内模拟甲烷氧化过程中,三种草地土壤中甲烷氧化菌数量均显著增加,达到3.26×107 g/d.w.s.到5.98×107 g/d.w.s (图 1-C)。相关性分析表明,三种类型草地土甲烷氧化潜力与原位甲烷氧化菌的pmoA基因拷贝丰度具有正相关关系(R2 = 0.5537) (图 1-D)。
 |
| 图 1 高浓度CH4培养(10000 μL/L)下三种草地土壤的甲烷氧化动力学过程(A)、甲烷氧化潜力(B)、甲烷氧化菌pmoA基因丰度(C)以及甲烷氧化潜力(MOP)与背景土壤中pmoA基因拷贝丰度之间的相关性分析(D) Figure 1 Methane oxidation kinetics (A), methane oxidation potential (B), methane-oxidizing bacteria abundance (C), relationship between methane oxidation potential (MOP) and pmoA gene copy number in Day 0 soil (D) of three grassland soils incubation at 10000 μL/L CH4. Day 0: In situ soil before incubation; Day 8: Soil after 8 days incubation |
| 图选项 |
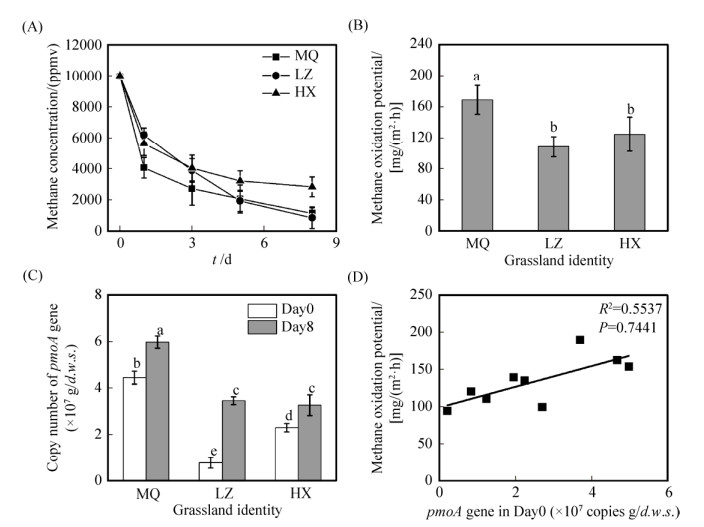
三种草地土壤中,原位甲烷氧化菌的组成也表现出明显的分异(图 2)。甲烷氧化菌pmoA基因的高通量测序分析结果表明,三种草地土壤中主要包括尚未被分离的难培养大气甲烷氧化菌、传统可培养TypeⅠ型和TypeⅡ型甲烷氧化菌,但是三类甲烷氧化菌的相对丰度却未表现出明显的差异;MQ、LZ和HX三种土壤中,TypeⅠ型甲烷氧化菌的相对丰度占比分别约为0.37%–23.27%、1.1%–57.37%和0.27%–0.32%;TypeⅡ型甲烷氧化菌的相对丰度占比分别约0.00%、0.37%–1.15%、0.08%–0.15%;难培养的大气甲烷氧化菌的相对丰度占比分别约8.46%–44.55%、15.36%–17.36%、79.25%–85.62%;值得注意的是,生物重复之间具有较大的空间异质性,表明不同微环境也可能选择了特定的微生物类群。进一步的分析则表明:在原位土壤中,HX土壤中难培养的大气甲烷氧化菌占主导,特别是USCγ,占比高达90%左右。LZ和MQ土壤具有较大的空间异质性,其中L-1和L-2中优势类群为USCγ和JR3,其中的JR3占比均高于60%;M-1和M-2中优势菌群为RA21和USCγ,但在LZ (L-3)和MQ (M-3)样品中以经典的TypeⅠ甲烷氧化菌(Methylosarcina)占主导,其占比为78%。
 |
| 图 2 高浓度CH4培养(10000 μL/L)下三种草地土壤(每种土壤包含3个生物重复)的甲烷氧化菌群落组成的变化规律 Figure 2 The population dynamics of methane oxidizing bacteria in three grassland soils (each soil consists of three biological repetitions) during consumption of methane at a concentration of 10000 μL/L for 8 days |
| 图选项 |
室内模拟高浓度甲烷氧化过程则得到明显不同的结果。高浓度甲烷培养8 d后,三种土壤中TypeⅠ、TypeⅡ型的传统甲烷氧化菌相对丰度显著增加,HX土壤中MOB群落的变化较小,优势类群仍是USCγ;LZ土壤的3个生物重复中,均为TypeⅠ型的Methylosarcina的相对丰度增加,成为绝对优势种,占比均达到了67%以上,与原位(Day0)相比,分别增加了15.3倍、8.92倍、1.25倍,表明在LZ土壤中氧化高浓度甲烷的微生物可能是TypeⅠ的Methylosarcina发挥了重要作用;MQ土壤中,M-1和M-2中TypeⅠ型的Methylosarcina发生明显增加,分别增加了10.72倍和5.91倍,而在M-3中主要是TypeⅡ型的Methylocystis和pmoA-2大量增加,幅度达28%以上。
本研究发现三种典型草地土壤中均以难培养的大气甲烷氧化菌为优势类群(图 2),与前期野外大气甲烷氧化通量观测结果吻合,如在甘肃环县天然草地[26]甲烷氧化最大和最小速率分别为0.25 CH4 mg/(m2·min)和0.14 CH4 mg/(m2·min),临泽(西北内陆干旱区)人工草地[27]生长季内大气CH4氧化量呈现先降低后升高的趋势,而青藏高原玛曲[28]连续两年高寒草甸非生长季吸收CH4,非生长季CH4累积吸收通量对全年累积通量的贡献为45.5%。针对这些典型草原类型,通过高通量测序土壤中甲烷氧化菌pmoA基因,结果表明难培养大气甲烷氧化菌为优势类群,可能在草地土壤原位甲烷氧化过程中发挥了重要作用。例如,环县HX土壤中USCγ为优势类群,占比高达90%;而USCγ和JR3是临泽和玛曲草地土壤中的优势类群,占比分别高达80%以上。Pravin等[29]也得到了类似结果,通过原位监测和分子生态学技术,发现德国吉森草地土壤不仅具有大气甲烷氧化能力,而且USCγ是主要的大气甲烷氧化菌。我国草地土壤研究也得到相似结果,Zheng等[30]采用120 μL/L甲烷培养2周,发现青藏高原海北高山草甸土壤可氧化88%的甲烷,进一步构建克隆文库发现USCγ是优势的大气甲烷氧化菌,占比高达98.4%,此外,研究者也发现了土壤中存在传统可培养TypeⅠ和TypeⅡ等甲烷氧化菌。但是,目前尚未有研究能够在土壤大气甲烷氧化过程及其微生物作用者之间建立直接的联系,草地土壤中传统可培养甲烷氧化菌的功能及其在原位甲烷氧化过程中的作用仍需进一步研究。
值得注意的是,草地土壤中既含有大气甲烷氧化菌,也含有传统甲烷氧化菌,意味着草地土壤具有高浓度甲烷氧化的潜力,但目前相关研究报道较少。我们采用高浓度甲烷培养,发现草地土壤具有低亲和力甲烷氧化能力,其中的传统甲烷氧化菌可能在10000 μL/L高浓度甲烷氧化过程中发挥了重要作用。培养8 d之后,LZ草地土壤TypeⅠ型的Methylosarcina和Methylocaldum 2个属的相对丰度均明显增加,成为高浓度甲烷下绝对优势类群,占比高达67%以上,而MQ土壤中TypeⅠ谱系的Methylosarcina属、TypeⅡ谱系的Methylocystis和pmoA-2类群也发生了显著增加。由于动物放牧行为造成地上植物群落组成和地下生物地球化学特性的差异[31],导致平行样点间存在一定的空间异质性,未表现出统计上的显著性。本研究中甘肃省的3个不同生态区的地理位置及基本情况如表 1所示,差异显著。但整体而言,高浓度CH4培养后TypeⅠ和TypeⅡ甲烷氧化菌明显成为LZ和MQ草地土壤的优势类群,表明这些微生物极可能在高浓度甲烷氧化过程中发挥了重要作用。值得注意的是,高浓度甲烷培养环县HX土壤8 d后,尽管Type Ⅰ型的Methylosarcina和Methylocaldum均发生明显富集,但难培养大气甲烷氧化菌仍是优势微生物类群。这些结果表明草地土壤也可能经历高浓度甲烷环境,促进低亲和力甲烷氧化菌的生长。事实上,草原雨季中土壤水分含量会急剧增加,导致氧气浓度减少并形成厌氧微域,进而导致产甲烷古菌活性增强释放出大量甲烷。而随着水分的减少,高浓度甲烷可能作为底物,显著刺激了传统甲烷氧化菌的生长。未来仍需开展更多的原位甲烷浓度观测实验,结合室内分子生态学研究,解析草地土壤甲烷氧化微生物过程的环境驱动机制。
土壤理化性质是影响甲烷氧化微生物生理活性的重要因子。pH影响大气甲烷氧化菌的地理分异规律,USCα通常存在于酸性土壤,而USCγ主要存在于pH > 6的土壤[32]。本研究使用的所有土壤的pH均高于6。同时,我们在所有三种土壤中发现的难培养大气甲烷氧化菌绝大部分属于USCγ类群。土壤无机氮也被认为对甲烷氧化菌具有重要影响。有研究发现土壤中无机氮可以通过抑制大气甲烷氧化菌的生长或通过抑制其甲烷氧化酶活性来影响大气甲烷的吸收[33]。本研究中,MQ的土壤无机氮含量较高,原位观测实验也发现MQ草地土壤的甲烷通量较高。此外,植被类型也可能对甲烷吸收产生很大影响[34-35]。Maurer等认为植物根系和凋落物可能产生高浓度单萜类物质,而这些物质可能被细菌矿化,对pMMO有抑制作用从而影响甲烷的氧化过程[36]。本研究的三种草地的优势植物种类不同(表 1),可能在一定程度上影响了三种草地的甲烷氧化菌的群落结构及其甲烷氧化潜力,但具体植物及其根系分泌物和凋落物如何影响高亲和力和低亲和力甲烷氧化,未来仍需通过组学和示踪技术,准确鉴定草地土壤中的活性甲烷氧化菌及其环境调控机制。
3 结论 利用高通量测序技术,通过原位分析和室内微宇宙培养,研究了三种类型草地土壤甲烷氧化菌丰度、多样性及其与甲烷氧化潜力之间的关系,发现高亲和力的难培养大气甲烷氧化菌是草地土壤中的优势类群,而草地土壤中传统可培养甲烷氧化菌在高浓度甲烷氧化过程中可能发挥了重要作用。这些结果为准确评估我国草地生态系统甲烷汇提供了基础数据,并为相关的微生物机制研究提供参考。未来可采用先进的土壤组学技术和稳定性同位素示踪技术,深入解析草地土壤中不同生理代谢特征的好氧甲烷氧化微生物过程。
References
| [1] | Dlugokencky EJ. Trends in Atmospheric Methane. NOAA/ESRL. 2019-04-05 (www.esrl.noaa.gov/gmd/ccgg/trends_ch4/) |
| [2] | Saunois M, Jackson RB, Bousquet P, Poulter B, Canadell JG. The growing role of methane in anthropogenic climate change. Environmental Research Letters, 2016, 11(12): 120207. DOI:10.1088/1748-9326/11/12/120207 |
| [3] | Topp E, Pattey E. Soils as sources and sinks for atmospheric methane. Canadian Journal of Soil Science, 1997, 77(2): 167-177. DOI:10.4141/S96-107 |
| [4] | Hartmann AA, Buchmann N, Niklaus PA. A study of soil methane sink regulation in two grasslands exposed to drought and N fertilization. Plant and Soil, 2011, 342(1/2): 265-275. |
| [5] | Unwin D. Climate change 1995: the science of climate change: JT Houghton, LG Meira Filho, BA Callander, N Harris, N Kattenberg, K Maskell (eds). Cambridge University Press, Cambridge (1996), 572 pp. £65.00 hardback: £22.95 paperback. Applied Geography, 1997, 17(2): 163. |
| [6] | Wang YF, Chen H, Zhu QA, Peng CH, Wu N, Yang G, Zhu D, Tian JQ, Tian LX, Kang XM, He YX, Gao YH, Zhao XQ. Soil methane uptake by grasslands and forests in China. Soil Biology and Biochemistry, 2014, 74: 70-81. DOI:10.1016/j.soilbio.2014.02.023 |
| [7] | Ishizuka S, Sakata T, Ishizuka K. Methane oxidation in Japanese forest soils. Soil Biology and Biochemistry, 2000, 32(6): 769-777. DOI:10.1016/S0038-0717(99)00200-X |
| [8] | Nanba K, King GM. Response of atmospheric methane consumption by maine forest soils to exogenous aluminum salts. Applied and Environmental Microbiology, 2000, 66(9): 3674-3679. DOI:10.1128/AEM.66.9.3674-3679.2000 |
| [9] | Schnell S, King GM. Mechanistic analysis of ammonium inhibition of atmospheric methane consumption in forest soils. Applied and Environmental Microbiology, 1994, 60(10): 3514-3521. |
| [10] | Hu MJ, Tong C, Zou FF. Effects of nitrogen input on CH4 production, oxidation and transport in soils, and mechanisms: a review. Acta Prataculturae Sinica, 2015, 24(6): 204-212. (in Chinese) 胡敏杰, 仝川, 邹芳芳. 氮输入对土壤甲烷产生、氧化和传输过程的影响及其机制. 草业学报, 2015, 24(6): 204-212. |
| [11] | Ma G, Wang P, Wang DX, Xu SQ. Response of soil greenhouse gas emissions to different forms of nitrogen in alpine shrub ecosystems. Acta Prataculturae Sinica, 2015, 24(3): 20-29. (in Chinese) 马钢, 王平, 王冬雪, 徐世权. 高寒灌丛土壤温室气体释放对添加不同形态氮素的响应. 草业学报, 2015, 24(3): 20-29. |
| [12] | Zhou XQ, Wang YF, Huang XZ, Tian JQ, Hao YB. Effect of grazing intensities on the activity and community structure of methane-oxidizing bacteria of grassland soil in Inner Mongolia. Nutrient Cycling in Agroecosystems, 2008, 80(2): 145-152. DOI:10.1007/s10705-007-9127-1 |
| [13] | Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Frontiers in Microbiology, 2015, 6: 1346. |
| [14] | Costello AM, Lidstrom ME. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Applied and Environmental Microbiology, 1999, 65(11): 5066-5074. |
| [15] | Steudler PA, Bowden RD, Melillo JM, Aber JD. Influence of nitrogen fertilization on methane uptake in temperate forest soils. Nature, 1989, 341(6240): 314-316. DOI:10.1038/341314a0 |
| [16] | Striegl RG, McConnaughey TA, Thorstenson DC, Weeks EP, Woodward JC. Consumption of atmospheric methane by desert soils. Nature, 1992, 357(6374): 145-147. DOI:10.1038/357145a0 |
| [17] | Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiology Letters, 1995, 132(3): 203-208. DOI:10.1111/fml.1995.132.issue-3 |
| [18] | Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen K, Murrell JC. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Applied and Environmental Microbiology, 1999, 65(8): 3312-3318. |
| [19] | Knief C, Lipski A, Dunfield PF. Diversity and activity of methanotrophic bacteria in different upland soils. Applied and Environmental Microbiology, 2003, 69(11): 6703-6714. DOI:10.1128/AEM.69.11.6703-6714.2003 |
| [20] | Ma TL, Chen H, Wang YF, Kang XM, Tian JP, Zhou XQ, Zhu QA, Peng CH, Liu LF, Hu J, Zhan W, Zhu EX. Effects of enclosure time on the community composition of methanotrophs in the soils of the Inner Mongolia grasslands. Journal of Soils and Sediments, 2016, 16(3): 1022-1031. DOI:10.1007/s11368-015-1305-1 |
| [21] | Kou YP, Li JB, Wang YS, Li CN, Tu B, Yao MJ, Li XZ. Scale-dependent key drivers controlling methane oxidation potential in Chinese grassland soils. Soil Biology and Biochemistry, 2017, 111: 104-114. DOI:10.1016/j.soilbio.2017.04.005 |
| [22] | Zheng Y, Jia ZJ. Next generation sequencing and stable isotope probing of active microorganisms responsible for aerobic methane oxidation in red paddy soils. Acta Microbiologica Sinica, 2013, 53(2): 173-184. (in Chinese) 郑燕, 贾仲君. 新一代高通量测序与稳定性同位素示踪DNA/RNA技术研究稻田红壤甲烷氧化的微生物过程. 微生物学报, 2013, 53(2): 173-184. |
| [23] | Holm PE, Nielsen PH, Albrechtsen HJ, Christensen TH. Importance of unattached bacteria and bacteria attached to sediment in determining potentials for degradation of xenobiotic organic contaminants in an aerobic aquifer. Applied and Environmental Microbiology, 1992, 58(9): 2016-2025. |
| [24] | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 2007, 73(16): 5261-5267. DOI:10.1128/AEM.00062-07 |
| [25] | Cai YF, Zheng Y, Bodelier PLE, Conrad R, Jia ZJ. Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nature Communications, 2016, 7: 11728. DOI:10.1038/ncomms11728 |
| [26] | Yang HL, Chen XJ, Hou FJ. Greenhouse gases emissions of rangeland and livestock manure in the eastern Gansu Loess Plateau in summer. Pratacultural Science, 2016, 33(8): 1454-1459. (in Chinese) 杨晗蕾, 陈先江, 侯扶江. 陇东黄土高原草地与畜粪夏季的温室气体排放. 草业科学, 2016, 33(8): 1454-1459. |
| [27] | Ning J. Greenhouse gas emission from growing season of sown pastures in inland arid area. Master Dissertation of Lanzhou University, 2016. (in Chinese) 宁娇.内陆干旱区栽培草地生长季温室气体排放.兰州大学硕士学位论文, 2016. |
| [28] | Liu Y, Yan CY, Matthew C, Wood B, Hou FJ. Key sources and seasonal dynamics of greenhouse gas fluxes from yak grazing systems on the Qinghai-Tibetan Plateau. Scientific Reports, 2017, 7: 40857. DOI:10.1038/srep40857 |
| [29] | Shrestha PM, Kammann C, Lenhart K, Dam B, Liesack W. Linking activity, composition and seasonal dynamics of atmospheric methane oxidizers in a meadow soil. The ISME Journal, 2012, 6(6): 1115-1126. DOI:10.1038/ismej.2011.179 |
| [30] | Zheng Y, Yang W, Sun X, Wang SP, Rui YC, Luo CY, Guo LD. Methanotrophic community structure and activity under warming and grazing of alpine meadow on the Tibetan Plateau. Applied Microbiology and Biotechnology, 2012, 93(5): 2193-2203. DOI:10.1007/s00253-011-3535-5 |
| [31] | Liu C. The mechanisms by grazing on grassland plant and soil spatial heterogeneity and their correlation. Doctor Dissertation of Northeast Normal University, 2015. (in Chinese) 刘晨.放牧对草地植被、土壤空间异质性及其相互关系的调控机制.东北师范大学博士学位论文, 2015. |
| [32] | Kolb S, Knief C, Dunfield PF, Conrad R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environmental Microbiology, 2005, 7(8): 1150-1161. DOI:10.1111/emi.2005.7.issue-8 |
| [33] | Aronson EL, Dubinsky EA, Helliker BR. Effects of nitrogen addition on soil microbial diversity and methane cycling capacity depend on drainage conditions in a pine forest soil. Soil Biology and Biochemistry, 2013, 62: 119-128. DOI:10.1016/j.soilbio.2013.03.005 |
| [34] | Borken W, Xu YJ, Beese F. Conversion of hardwood forests to spruce and pine plantations strongly reduced soil methane sink in Germany. Global Change Biology, 2003, 9(6): 956-966. DOI:10.1046/j.1365-2486.2003.00631.x |
| [35] | Li CN, Li JB, Li XZ. Soil methanotrophic community structure and diversity in different vegetation types at elevation gradient of Gongga Mountain, Southwest China. Chinese Journal of Applied Ecology, 2017, 28(3): 805-814. (in Chinese) 李超男, 李家宝, 李香真. 贡嘎山海拔梯度上不同植被类型土壤甲烷氧化菌群落结构及多样性. 应用生态学报, 2017, 28(3): 805-814. |
| [36] | Maurer D, Kolb S, Haumaier L, Borken W. Inhibition of atmospheric methane oxidation by monoterpenes in Norway spruce and European beech soils. Soil Biology and Biochemistry, 2008, 40(12): 3014-3020. DOI:10.1016/j.soilbio.2008.08.023 |
