蒲秀鑫1,2, 柴丽娟2, 徐鹏翔1, 张晓娟2,4, 陆震鸣2,4, 王松涛4, 沈才洪4, 史劲松3, 许正宏1,2,4


1. 江南大学工业生物技术教育部重点实验室, 生物工程学院, 江苏 无锡 214122;
2. 江南大学粮食发酵工艺与技术国家工程实验室, 江苏 无锡 214122;
3. 江南大学药学院, 江苏 无锡 214122;
4. 国家固态酿造工程技术研究中心, 四川 泸州 646000
收稿日期:2019-02-26;修回日期:2019-05-15;网络出版日期:2019-10-17
基金项目:国家自然科学基金(31771967,31901658);国家轻工技术与工程一流学科自主课题(2018-11)
*通信作者:许正宏, Tel/Fax:+86-510-85918206, E-mail:zhenghxu@jiangnan.edu.cn.
摘要:[目的] 分离窖泥中的梭菌微生物并对其代谢产物进行评估。[方法] 对窖泥中梭菌群落的16S rRNA基因进行高通量测序;利用高丰度的梭菌OTU序列在KOMODO数据库进行培养基的预测,定向分离窖泥中梭菌菌株;采用顶空固相微萃取结合气相色谱质谱联用仪对窖泥和代表性梭菌菌株的挥发性代谢产物进行检测。[结果] 利用KOMODO数据库预测的梭菌培养基共计筛选到31株梭菌微生物,分属于梭菌属的14个种;根据风味代谢特性,这些菌株主要分为两大类,一是C.carboxidivorans、C.sporogenes和C.tyrobutyricum等产酸为主的梭菌,二是C.beijerinckii、C.butyricum和C.sphenoides等产醇为主的梭菌。[结论] 利用测序序列预测培养基有助于从窖泥中分离获得丰富的梭菌菌株,其物种和代谢能力的多样性对解析白酒复杂风味形成机理奠定了一定的基础。
关键词:泸型酒窖泥梭菌KOMODO
Isolation and metabolic characters of Clostridium strains from pit mud of Luzhou-flavor baijiu
Xiuxin Pu1,2, Lijuan Chai2, Pengxiang Xu1, Xiaojuan Zhang2,4, Zhenming Lu2,4, Songtao Wang4, Caihong Shen4, Jinsong Shi3, Zhenghong Xu1,2,4


1. Key Laboratory of Industrial Biotechnology of Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu Province, China;
2. National Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi 214122, Jiangsu Province, China;
3. School of Pharmaceutical Sciences, Jiangnan University, Wuxi 214122, Jiangsu Province, China;
4. National Engineering Research Center of Solid-State Brewing, Luzhou 646000, Sichuan Province, China
Received: 26 February 2019; Revised: 15 May 2019; Published online: 17 October 2019
*Corresponding author: Zhenghong Xu, Tel/Fax:+86-510-85918206, E-mail:zhenghxu@jiangnan.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31771967, 31901658) and by the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE 2018-11)
Abstract: [Objective] This study aims to isolate Clostridium strains from Luzhou-flavor baijiu pit mud and evaluate their metabolic characteristics. [Methods] 16S rRNA gene sequencing was used to analyze the clostridial community structure in pit mud; specific media were predicted according to the sequences of high-abundance OTUs scattered in Clostridia using KOMODO database; volatile compounds of pit mud and the representative isolated Clostridium strains' fermentation broth were detected by headspace solid phase microextraction combined with gas chromatography-mass spectrometry. [Results] Thirty-one strains belonging to 14 species of genus Clostridium were isolated using the 7 media predicted by KOMODO database. Based on the analysis of volatile metabolites, these Clostridium strains could be divided into two clusters, including cluster I, i.e. strains mainly producing acids (C. carboxidivorans, C. sporogenes and C. tyrobutyricum) and cluster II, i.e. strains mainly producing alcohols (C. beijerinckii, C. butyricum and C. sphenoides). [Conclusion] KOMODO method is helpful for the isolation of Clostridium or other bacteria from the pit mud of Luzhou-flavor baijiu, and elucidating their metabolic profiles could be conductive to analyze the function of brewing microbes in baijiu flavor formation.
Keywords: Luzhou-flavor baijiupit mudClostridiumKOMODO
白酒是我国传统发酵型蒸馏酒[1],根据其风味特点可分为不同香型[2]。其中,浓香型白酒的产量占白酒总产量70%以上[3]。浓香型白酒是以高粱为主要原料,大曲为发酵剂,泥窖为发酵容器,经过多菌种混合发酵、固态蒸馏、勾调、陈酿等工艺酿造而成,具有窖香浓郁、绵甜醇厚、香味协调、回味悠长的特点[4]。
生产实践经验告诉我们“好窖出好酒”,说明了窖泥在浓香型白酒发酵形成风味过程中扮演了重要角色[5-6]。近年来随着微生物免培养技术的不断发展,已经分析了窖泥中存在的梭菌、芽孢杆菌、甲烷杆菌等种类复杂的微生物多样性[7-8]。其中,梭菌是优质窖泥中的优势微生物,对风味物质形成具有重要影响。例如,C. carboxidivorans[9]、C. beijerinckii[10]、C. butyricum[11]和C. tyrobutyricum[12-13]等梭菌被证实具有产生乙酸、丁酸、己酸和丁醇等小分子化合物的能力。但是,目前窖泥中仍然有许多梭菌尚未纯培养分离,对其在窖泥中的代谢功能尚缺乏深入了解。
本研究首先对窖泥中细菌群落进行高通量测序,根据基因序列的丰度和亲缘关系获得梭菌代表性OTU;其次利用OTU序列在KOMODO[14]数据库(http://komodo.modelseed.org/)中进行梭菌培养基的预测,定向分离窖泥梭菌菌株;最后,采用顶空固相微萃取结合气相色谱质谱联用仪对代表性梭菌菌株的代谢产物进行检测。本研究为明确窖泥中梭菌群落的结构和功能,解析窖泥风味来源和白酒酿造机理奠定了研究基础。
1 材料和方法 1.1 材料
1.1.1 样品采集: 取样地点为泸州老窖股份有限公司酿酒基地的新窖窖池(约30年)和老窖窖池(约200年)。在窖池开封后进行底部三点取样,每个点约5 g窖泥,放入无菌塑封袋后尽快用于微生物分离。
1.1.2 主要试剂和仪器: PowerSoil DNA提取试剂盒,美国MOBIO公司;细菌基因组DNA提取试剂盒,上海捷瑞生物工程有限公司;2-辛醇,上海阿拉丁生化科技股份有限公司;气相色谱质谱联用仪Trace1310 ISQ和NanoDrop 2000超微量分光光度计,美国赛默飞世尔科技公司。
1.2 窖泥宏基因组提取 采用PowerSoil DNA提取试剂盒对窖泥宏基因组进行提取,步骤参考说明书。用超微量分光光度计测定DNA的浓度和纯度。保证样品DNA质量大于5 μg且A260/A280值为1.8-2.0[15]。
1.3 细菌16S rRNA基因测序 采用细菌16S rRNA基因V3-V4区通用引物338F (5′-ACTCCTACGGGAGGCAGCAG-3′)与806R (5′-GGACTACHVGGGTWTCTAAT-3′),合成带有6 bp碱基Barcode的特异性引物,对样本进行16S高通量测序。测序平台为Illumina MiSeq,按照测序标准流程进行测序上机操作,由华大基因股份有限公司完成。
1.4 梭菌培养基的预测 根据细菌高通量测序结果中梭菌纲OTU的序列条目,构建系统发育树,结合梭菌纲OTU的丰度和亲缘关系,找出代表性梭菌OTU序列,将所得到的OTU序列在KOMODO网站[14]上进行培养基种类的预测。
1.5 梭菌的分离 根据KOMODO预测的结果,采用德国微生物菌种保藏中心(DSMZ)数据库中的Medium 1、9、21、78、339、634和693这7种培养基对窖泥中梭菌进行分离培养。
1.6 提取细菌DNA 采用细菌基因组DNA提取试剂盒对细菌的DNA进行提取,步骤参考说明书。并利用NanoDrop 2000超微量分光光度计检测DNA的质量。
1.7 16S rRNA基因扩增及菌种鉴定 采用PCR扩增仪对基因组进行扩增。所采用的引物为细菌通用引物:27F (5′-AGAGTTTGATC CTGGCTCAG-3′)与1492R (5′-GGTTACCTTGTTA CGACTT-3′)。PCR反应体系为(25 μL):ExTaq酶12.5 μL,引物27F 1 μL,引物1492R 1 μL,模板DNA 1 μL,ddH2O 9.5 μL。PCR反应程序为:95 ℃ 4 min;95 ℃ 1 min,56 ℃ 1 min,72 ℃ 90 s,共30个循环;终延伸72 ℃ 10 min。
PCR产物使用1.2%的琼脂糖凝胶电泳检验后,送至无锡天霖测序公司测序,将所得到的结果提交至GenBank进行在线BLAST比对分析,进行物种鉴定。
1.8 风味物质检测
1.8.1 样品预处理: (1) 发酵液处理方法:选取代表性菌株在对应培养基中活化至对数期后转接于液体发酵培养基中,置于厌氧培养箱培养7 d,吸取菌株培养液10 mL至离心管中,离心取8 mL上清至萃取瓶中,加入2.8 g氯化钠,并同时加入10 μL浓度为2.2 g/L的2-辛醇标准品用于分析。(2)窖泥处理方法:准确称取2 g窖泥于萃取瓶中,加入8 mL盐溶液(0.85% NaCl,1% CaCl2),然后加入2.8 g氯化钠,并同时加入10 μL浓度为2.2 g/L的2-辛醇标准品用于分析。
1.8.2 气相色谱及质谱检测条件[11]: (1) 萃取方法:采用美国Supelco公司的50 μm/30 μm DVAB/CAR/PDMS固相微萃取头,于50 ℃萃取30 min,将萃取后的固相萃取头插入气相色谱仪进样口进行热解吸附5 min,进样口温度250 ℃。(2) GC-MS检测条件:色谱柱规格为Agilent DB-Wax毛细管柱(30 m×0.32 mm×0.25 μm),载气为氦气,流速为10 mL/min,不分流,进样口温度维持在250 ℃,升温程序为:40 ℃维持5 min之后,以5 ℃/min速率升温至60 ℃,以10 ℃/min速率升温至230 ℃,维持8 min。质谱条件:EI电离源,离子源温度为220 ℃,接口温度为250 ℃,离子源能量为70 eV,扫描范围:33-450 amu。
1.8.3 定性与半定量: 在NIST谱库和Willey谱库中检索出匹配度大于800的挥发性化合物,根据内标2-辛醇与风味物质峰面积之比计算出挥发性风味物质含量,再乘以稀释倍数得出样品中挥发性风味物质的含量。
1.9 数据分析 采用SPSS等软件进行数据统计分析,系统发育树构建采用MEGA 7,热图分析等采用Graphpad Prism 7.0、R语言等软件处理。
2 结果和分析 2.1 窖泥中微生物群落结构及代表性梭菌系统发育分析 对新窖窖泥和老窖窖泥中细菌的微生物群落结构从纲水平进行分析,结果如图 1-A所示。新窖泥细菌群落中主要是梭菌纲(Clostridia)、拟杆菌纲(Bacteroidia)和芽孢杆菌纲(Bacilli),相对丰度分别为54%、35%和7%;老窖泥细菌群落中梭菌纲和拟杆菌纲的相对丰度分别为80%和13%,而芽孢杆菌纲仅占0.29%。上述结果与Hu等[12]、邓杰[16]、任聪等[17]、罗雯等[18]的报道相似。从梭菌纲中筛选出相对丰度排名前55的代表性OTU序列,构建系统发育树,发现其分为9个类群(图 1-B)。
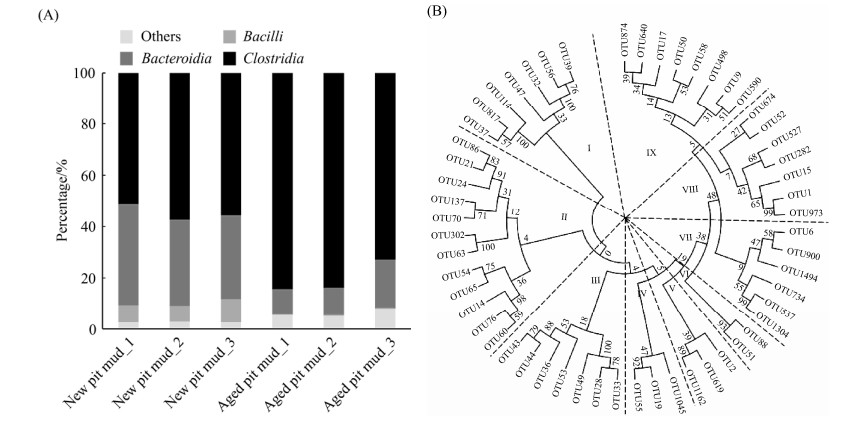 |
| 图 1 新老窖泥细菌群落结构(A)及基于邻接法构建的相对丰度前55的梭菌纲OTU序列系统发育树(B) Figure 1 The microbial community structure of new- and aged-pit mud at class level (A) and the phylogenetic tree of 55 representative Clostridial OTU sequences constructed by neighbor-joining method (B). |
| 图选项 |
2.2 窖泥中梭菌培养基的预测 将55条代表性OTU上传至KOMODO网站,筛选最高分值或得分在800以上的结果,总共获得了7种梭菌培养基:包括Medium 1、9、21、78、339、634和693,培养基条件参见德国微生物菌种保藏中心(DSMZ)数据库(表 1)。
表 1. 基于55条梭菌OTU序列的培养基预测结果 Table 1. Predicted media based on 55 Clostridial OTU sequences
| Classification | OTU number | Predicted medium | Score |
| Group Ⅰ | OTU 37 | Medium 1(DSMZ) | 1200 |
| OTU 39 | Medium 697(DSMZ) | 665 | |
| OTU 47 | Medium 971(DSMZ) | 400 | |
| OTU 114 | Medium 507(DSMZ) | 400 | |
| OTU 56,OTU 32,OTU 817 | No results found | ||
| Group Ⅱ | OTU 24 | Medium 9(DSMZ) | 150139 |
| OTU 21 | Medium 220(DSMZ) | 400 | |
| OTU 137 | Medium 58(DSMZ) | 400 | |
| OTU 63 | Medium 769(DSMZ) | 400 | |
| OTU 65 | Medium 53(DSMZ) | 400 | |
| OTU 76 | Medium 515(DSMZ) | 400 | |
| OTU 86,OTU 70,OTU 302,OTU 54,OTU 14,OTU 60 | No results found | ||
| Group Ⅲ | OTU 28 | Medium 693(DSMZ) | 800 |
| OTU 43 | Medium 220(DSMZ) | 400 | |
| OTU 44 | Medium 339(DSMZ) | 400 | |
| OTU 36 | Medium 788(DSMZ) | 400 | |
| OTU 53 | Medium 92(DSMZ) | 400 | |
| OTU 49 | Medium 436(DSMZ) | 400 | |
| OTU 33 | No results found | ||
| Group Ⅳ | OTU 19 | Medium 220(DSMZ) | 400 |
| OTU 55,OTU 1045, | No results found | ||
| Group Ⅴ | OTU 1162,OTU 619,OTU 2 | No results found | |
| Group Ⅵ | OTU 88 | Medium 65(DSMZ) | 400 |
| OTU 51 | Medium 156(DSMZ) | 400 | |
| Group Ⅶ | OTU 1304 | Medium 104(DSMZ) | 400 |
| OTU 1494 | Medium 123(DSMZ) | 400 | |
| OTU 900 | Medium 1237(DSMZ) | 400 | |
| OTU 6 | Medium 110(DSMZ) | 400 | |
| OTU 537,OTU 734 | No results found | ||
| Group Ⅷ | OTU 674 | Medium 634(DSMZ) | 2400 |
| OTU 282 | Medium 78(DSMZ) | 2000 | |
| OTU 527 | Medium 339(DSMZ) | 800 | |
| OTU 52,OTU 973 | Medium 339(DSMZ) | 400 | |
| OTU 15 | Medium 104(DSMZ) | 400 | |
| OTU 1 | No results found | ||
| Group Ⅸ | OTU 874 | Medium 78(DSMZ) | 2000 |
| OTU 50 | Medium 21(DSMZ) | 1600 | |
| OTU 498,OTU 58 | Medium 110(DSMZ) | 400 | |
| OTU 590,OTU 9,OTU 17,OTU 640 | No results found |
表选项
2.3 窖泥中梭菌菌株的分离 从窖泥样品中分离获得31株梭菌微生物,分属于梭菌属的14个种(表 2)。
表 2. 基于14株代表性菌株16S rRNA序列的生物鉴定比对表 Table 2. BLAST results based on the 16S rRNA sequences of representative isolated Clostridium strains
| Number | Most similar strain | Similarity/% | Accession No. |
| C1 | C. senegalense | 100 | NR_125591.1 |
| C2 | C. sphenoides | 99 | NR_112174.1 |
| C3 | C. aminovalericum | 97 | NR_113199.1 |
| C4 | C. homopropionicum | 98 | NR_026148.1 |
| C5 | C. celerecrescens | 99 | NR_026100.1 |
| C6 | C. butyricum | 99 | NR_113244.1 |
| C7 | C. tepidum | 99 | NR_157639.1 |
| C8 | C. beijerinckii | 99 | NR_113388.1 |
| C9 | Clostridium sp. | 100 | AB610569.1 |
| C10 | C. botulinum | 99 | CP027775.1 |
| C11 | C. scatologenes | 99 | CP009933.1 |
| C12 | C. carboxidivorans | 99 | NR_104768.1 |
| C13 | C. sporogenes | 99 | NR_113245.1 |
| C14 | C. tyrobutyricum | 100 | NR_044718.2 |
表选项
将14个不同种梭菌菌株的16S rRNA基因序列与窖泥细菌群落高通量测序结果进行比对,分析不同梭菌在群落中的相对丰度(图 2)。C. tyrobutyricum在新窖泥和老窖泥中相对丰度较高,分别为0.16%和0.11%;C. sporogenes在老窖泥中平均相对丰度为0.21%,而在新窖泥中仅为0.03%。老窖泥中C. scatologenes的平均相对丰度为0.09%。
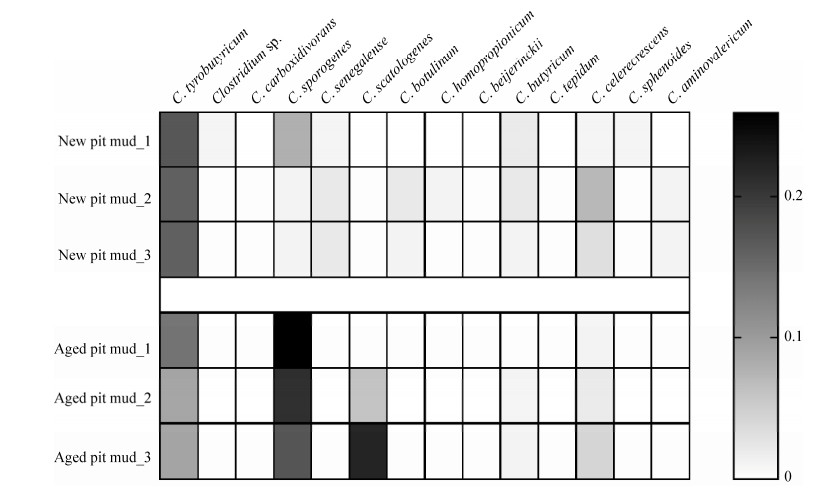 |
| 图 2 14株代表性梭菌在窖泥细菌群落中的相对丰度 Figure 2 Relative abundance of 14 representative Clostridium strains in pit mud. |
| 图选项 |
2.4 窖泥和梭菌发酵液中挥发性化合物分析 从窖泥中共检测出80种挥发性风味化合物,包括酯类45种,酸类9种,醇类10种,醛酮类9种以及7种其他化合物;梭菌发酵液中检测出239种化合物,其中酯类43种,酸类17种,醇类56种,醛酮类44种以及79种其他化合物。
窖泥中含量最多的为酯类物质,其次是酸和醇,而醛酮和其他物质含量很少(图 3)。窖泥A和B的酯类物质分别占总量的63%和76%,酸类物质分别占总量的30%和21%,醇类物质分别占总量的6%和2%。而梭菌的挥发性代谢物组成则随菌株的不同而具有明显的差异,但总体可归纳为:梭菌C1 (C. senegalense)、C2 (C. sphenoides)、C5 (C. celerecrescens)、C6 (C. butyricum)、C7 (C. tepidum)、C8 (C. beijerinckii)和C9 (Clostridium sp.)中醇类物质含量比较高。梭菌C3 (C. aminovalericum)、C4 (C. homopropionicum)、C11 (C. scatologenes)、C12 (C. carboxidivorans)、C13 (C. sporogenes)和C14 (C. tyrobutyricum)的发酵液中酸类物质含量比较高。梭菌代谢产生的酯类较少,除C9 (Clostridium sp.)外,其余梭菌试验条件下均没有表现出较突出的产酯能力。
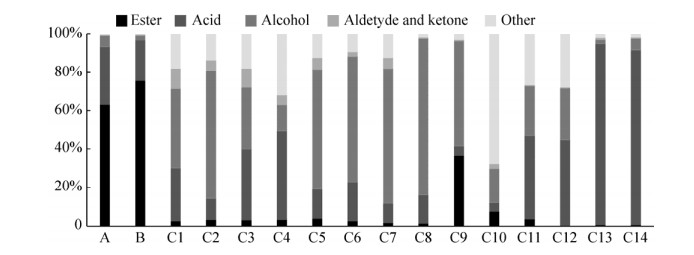 |
| 图 3 窖泥和梭菌发酵液的挥发性物质组成 Figure 3 Composition of volatile compounds in pit mud and the fermentation broth of isolated Clostridium strains. A: new pit mud; B: aged pit mud; C: strains. |
| 图选项 |
不同梭菌发酵液挥发性代谢产物的组成具有明显差异(图 4),梭菌C3 (C. aminovalericum)、C4(C. homopropionicum)、C11 (C. scatologenes)、C12 (C. carboxidivorans)、C13 (C. sporogenes)和C14 (C. tyrobutyricum)的发酵液中酸类物质含量比较高,主要为己酸、乙酸、丁酸和辛酸等。梭菌C1 (C. senegalense)、C2 (C. sphenoides)、C5 (C. celerecrescens)、C6 (C. butyricum)、C7 (C. tepidum)、C8 (C. beijerinckii)和C9 (Clostridium sp.)中醇类物质含量比较高,主要为己醇、丁醇和乙醇等。梭菌所产生的这些酸类物质和醇类物质在白酒发酵过程中可直接作为呈香物质,并且还可以在酶的催化作用下进一步形成相应的酯类化合物,如己酸乙酯、乙酸乙酯等,是白酒典型香气物质的主要来源。此外,梭菌还能代谢产生少量的醛酮类化合物、烷烃类化合物、吡嗪类化合物和一些其他物质,进一步影响白酒的品质。
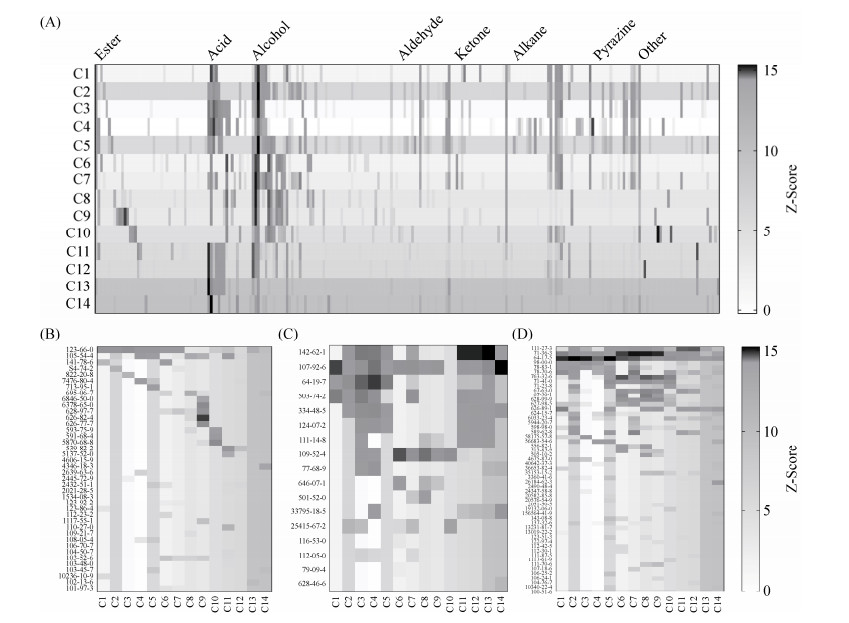 |
| 图 4 分离到的梭菌挥发性代谢产物热图分析 Figure 4 Heatmaps of volatile compounds of the isolated Clostridium strains' fermentation broth. A: Total volatiles; B: Esters; C: Acids; D: Alcohols. Cas numbers of the compounds were displayed on the left side of each subfigure. |
| 图选项 |
3 讨论 窖泥中的梭菌在白酒酿造过程中起到重要的功能和作用。任聪等发现老窖泥中的梭菌纲微生物占主导地位[17]。Hu等发现梭菌纲微生物在优质窖泥中的含量要高于其他品质的窖泥[19]。本研究利用高通量测序获得的梭菌OTU序列所预测的7个不同培养基,定向分离了窖泥中31个梭菌菌株,分属14个梭菌种,其中C. tyrobutyricum、C. butyricum、C. sporogenes、C. celerecrescens和C. carboxidivorans等梭菌在泸型酒窖泥中有分离报道[20-23],表明窖泥中这些梭菌较易分离培养,但对其代谢能力的研究较少。C. beijerinckii[10]、C. senegalense[24]、C. scatologenes[25]、C. sphenoides[26]、C. aminovalericum[27]、C. homopropionicum[28]、C. tepidum[29]等梭菌在白酒中未曾有过分离报道。通过纯培养实验验证了不同梭菌菌株可以代谢产生不同的白酒风味物质及其前体,其中C. sporogenes和C. tyrobutyricum具有较强的产己酸和丁酸的能力,丰富了我们对泸型酒梭菌群落组成和功能多样性的认识。
本文仅针对测序获得的高丰度梭菌OTU序列进行培养基预测与菌株分离,下一步需要对窖泥微生物群落中相对丰度较低的梭菌物种进行分离鉴定,评价其代谢特性,从而加强对窖泥中梭菌多样性和酿酒功能的了解。另外,本研究根据基因序列预测微生物培养基的方法也可能遇到没有办法准确预测,或者所预测的培养基仍然无法分离出目的菌株,这可能与现有的菌种库信息不完善有关。随着数据库和菌种库的数据不断更新,利用该方法预测培养基并分离目的菌株的准确度和可靠性将会进一步提高。
References
| [1] | Jin GY, Zhu Y, Xu Y. Mystery behind Chinese liquor fermentation. Trends in Food Science & Technology, 2017, 63: 18-28. |
| [2] | Zheng XW, Han BZ. Baijiu, Chinese liquor: History, classification and manufacture. Journal of Ethnic Foods, 2016, 3(1): 19-25. |
| [3] | Xu Y, Wang D, Fan WL, Mu XQ, Chen J. ChemInform abstract: traditional Chinese biotechnology. ChemInform, 2012, 43(50): 1522-2667. |
| [4] | Liu MK, Tang YM, Zhao K, Gu YF, Ren DQ, Yao WC, Tian XH, Liu Y, Zhang XP. Recent advances in research on the community, isolation, and application of microbes in the pit mud used in manufacture of Chinese Strong-flavor Baijiu. Microbiology China, 2017, 44(5): 1222-1229. (in Chinese) 刘茂柯, 唐玉明, 赵珂, 辜运富, 任道群, 姚万春, 田新惠, 刘颖, 张小平. 浓香型白酒窖泥微生物群落结构及其选育应用研究进展. 微生物学通报, 2017, 44(5): 1222-1229. |
| [5] | Wen CB, Li GH, Qiu SQ, Rao JQ, Du LQ. Study on the improvement of pit mud quality. Liquor-Making Science & Technology, 2009(4): 68-70. (in Chinese) 文成兵, 李光辉, 邱声强, 饶家权, 杜礼泉. 提高窖泥质量的研究. 酿酒科技, 2009(4): 68-70. |
| [6] | Zhang Q, Shen CH, Liu QB, Deng B, Ao ZH, Qian ZW, Liang ZX, Yuan H. Research progress in the evaluation of pit mud quality. Liquor-Making Science & Technology, 2013(7): 84-86. (in Chinese) 张强, 沈才洪, 刘清斌, 邓波, 敖宗华, 钱志伟, 梁泽新, 袁瀚. 窖泥质量评价研究进展. 酿酒科技, 2013(7): 84-86. |
| [7] | 胡晓龙.浓香型白酒窖泥中梭菌群落多样性与窖泥质量关联性研究.江南大学博士学位论文, 2015. http://cdmd.cnki.com.cn/Article/CDMD-10295-1016030626.htm |
| [8] | Tao Y, Li JB, Rui JP, Xu ZC, Zhou Y, Hu XH, Wang X, Liu MH, Li DP, Li XZ. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Applied and Environmental Microbiology, 2014, 80(7): 2254-2260. DOI:10.1128/AEM.04070-13 |
| [9] | Fernández-Naveira , Abubackar HN, Veiga MC, Kennes C. Efficient butanol-ethanol (B-E) production from carbon monoxide fermentation by Clostridium carboxidivorans. Applied Microbiology and Biotechnology, 2016, 100(7): 3361-3370. DOI:10.1007/s00253-015-7238-1 |
| [10] | Li HG, Luo W, Gu QY, Wang Q, Hu WJ, Yu XB. Acetone, butanol, and ethanol production from cane molasses using Clostridium beijerinckii mutant obtained by combined low-energy ion beam implantation and N-methyl-N-nitro- N-nitrosoguanidine induction. Bioresource Technology, 2013, 137: 254-260. DOI:10.1016/j.biortech.2013.03.084 |
| [11] | 马箭.泸型酒窖池丁酸菌群落结构初步解析.江南大学硕士学位论文, 2016. http://cdmd.cnki.com.cn/Article/CDMD-10295-1016265272.htm |
| [12] | Hu XL, Du H, Xu Y. Identification and quantification of the caproic acid-producing bacterium Clostridium kluyveri in the fermentation of pit mud used for Chinese strong-aroma type liquor production. International Journal of Food Microbiology, 2015, 214: 116-122. DOI:10.1016/j.ijfoodmicro.2015.07.032 |
| [13] | Lawson PA. The taxonomy of the genus Clostridium: current status and future perspectives. Microbiology China, 2016, 43(5): 1070-1074. |
| [14] | Oberhardt MA, Zarecki R, Gronow S, Lang E, Klenk HP, Gophna U, Ruppin E. Harnessing the landscape of microbial culture media to predict new organism-media pairings. Nature Communications, 2015, 6: 8493. DOI:10.1038/ncomms9493 |
| [15] | 栗连会.泸型酒酒醅中乳酸菌和乳酸降解菌的多样性和代谢特性.江南大学硕士学位论文, 2016. http://cdmd.cnki.com.cn/Article/CDMD-10295-1016265415.htm |
| [16] | 邓杰.基于高通量测序的浓香型白酒窖泥微生物群落结构研究.四川理工学院硕士学位论文, 2015. http://cdmd.cnki.com.cn/Article/CDMD-10622-1015972895.htm |
| [17] | Ren C, Gu Y, Du H, Xu Y. Predicting dominant caproate-producing microbes by comparing the microbiotas between new- and aged-pit muds. Food and Fermentation Industries, 2018, 44(12): 8-14. (in Chinese) 任聪, 辜杨, 杜海, 徐岩. 基于新老窖泥的微生物菌群结构判定浓香型白酒生产中的主体己酸菌. 食品与发酵工业, 2018, 44(12): 8-14. |
| [18] | Luo W, Zhang QY, Liao ZM, Zhang WX. Research on microbial composition of pit mud with different traits using high-throughput sequencing. Food and Fermentation Industries, 2017, 43(9): 9-14. (in Chinese) 罗雯, 张倩颖, 廖作敏, 张文学. 基于高通量测序技术的不同性状窖泥微生物组成研究. 食品与发酵工业, 2017, 43(9): 9-14. |
| [19] | Hu XL, Du H, Ren C, Xu Y. Illuminating anaerobic microbial community and cooccurrence patterns across a quality gradient in Chinese liquor fermentation pit muds. Applied and Environmental Microbiology, 2016, 82(8): 2506-2515. DOI:10.1128/AEM.03409-15 |
| [20] | Yang JJ, Tang X, Yang XC, Liu HJ, Tang YM, Chen Q. Genetic diversity and phylogeny of caproic acid bacteria in different aged Luzhou Laojiao Pit Mud. Journal of Sichuan Agricultural University, 2015, 33(2): 181-188. (in Chinese) 杨菁瑾, 唐雪, 杨先超, 刘汉军, 唐玉明, 陈强. 泸州老窖不同窖龄窖泥中己酸菌遗传多样性及系统发育. 四川农业大学学报, 2015, 33(2): 181-188. |
| [21] | Xue ZK, Xue Y. Polyphasic identification of a hexanoic acid bacteria strain from the pit mud and study on its acid producing ability. Liquor-Making Science & Technology, 2016(6): 65-71. (in Chinese) 薛正楷, 薛原. 一株窖泥己酸菌株的多相鉴定及产酸研究. 酿酒科技, 2016(6): 65-71. |
| [22] | He PX, Li FL, Zheng Y, Zhang Y, Hu XL, Sun XY, Guo FL. Isolation and volatile metabolites of Clostridium spp. in pit mud of strong-flavor Baijiu. China Brewing, 2017, 36(4): 45-49. (in Chinese) 何培新, 李芳莉, 郑燕, 张勇, 胡晓龙, 孙西玉, 郭福利. 浓香型白酒窖泥梭菌的分离及其挥发性代谢产物分析. 中国酿造, 2017, 36(4): 45-49. |
| [23] | Yuan HW, Zhao M, Jiang J, Liu GQ, Hu YQ, Shen CH, Wu ZD, Zhou RQ, Song C. Isolation and identification of a butyric acid-producing strain from pit mud and study on its growth performance. Liquor-Making Science & Technology, 2017(5): 35-40. (in Chinese) 袁华伟, 赵猛, 蒋君, 刘光钱, 胡永奇, 沈才洪, 吴重德, 周荣清, 宋川. 窖泥中产丁酸菌的筛选、鉴定及生长性能研究. 酿酒科技, 2017(5): 35-40. |
| [24] | Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Standards in Genomic Sciences, 2012, 6(3): 386-395. |
| [25] | Liu HX, Song TS, Fei KQ, Wang HQ, Xie JJ. Microbial electrosynthesis of organic chemicals from CO2 by Clostridium scatologenes ATCC 25775T. Bioresources and Bioprocessing, 2018, 5: 7. DOI:10.1186/s40643-018-0195-7 |
| [26] | Tran-Din K, Gottschalk G. Formation of D(-)-1, 2-propanediol and D(-)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Archives of Microbiology, 1985, 142(1): 87-92. |
| [27] | Barker HA, D'Ari L, Kahn J. Enzymatic reactions in the degradation of 5-aminovalerate by Clostridium aminovalericum. Journal of Biological Chemistry, 1987, 262(19): 8994-9003. |
| [28] | D?rner C, Schink B. Clostridium homopropionicum sp. nov., a new strict anaerobe growing with 2-, 3-, or 4-hydroxybutyrate. Archives of Microbiology, 1990, 154(4): 342-348. DOI:10.1007/BF00276529 |
| [29] | Dobritsa AP, Kutumbaka KK, Werner K, Wiedmann M, Asmus A, Samadpour M. Clostridium tepidum sp. nov., a close relative of Clostridium sporogenes and Clostridium botulinum Group Ⅰ. International Journal of Systematic and Evolutionary Microbiology, 2017, 67(7): 2317-2322. DOI:10.1099/ijsem.0.001948 |
