Wanli Peng, Mina Guli, Daning Zheng, Rubing Liang


State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China
Received: 26 October 2018; Revised: 19 December 2018; Published online: 13 March 2019
Foundation item: Supported by the National Natural Science Foundation of China (31570099, 31370152)
Corresponding author: Rubing Liang, Tel: +86-21-34204192; E-mail: icelike@sjtu.edu.cn.
Abstract: [Objective] Pseudomonas putida SJTE-1 can degrade 17β-estradiol (E2) efficiently, but the enzymes for the tranformation of E2 in this strain is still unclear. In this work, we identified and characterized a new 3-oxoacyl-acyl-carrier-protein reductase (3-oxoacyl-ACP reductase, ANI01589.1) involved in the E2 degradation.[Methods] We cloned the encoding gene of this 3-oxoacyl-ACP reductase and overexpressed it in Escherichia coli BL21(DE3) strain. We purified the recombinant protein by metal-ion affinity chromatography and characterized its enzymatic activity in vitro. Then we detected the product of this enzymatic reaction with High Performance Liquid Chromatography (HPLC).[Results] The transcription of the 3-oxoacyl-ACP reductase was induced by 17β-estradiol. Protein sequence alignment showed that it contained two consensus regions and the conserved residues of the short-chain dehydrogenase/reductase (SDR), and its structure was similar to that of the 3-oxoacyl-ACP reductase (4fw8.1.A). The recombinant protein was purified with the yield of 19.6 mg per liter culture. This 3-oxoacyl-ACP reductase could convert 17β-estradiol into estrone using NAD+ as the cofactor. Its Km value was 0.071 mmol/L and its kcat value was 2.4±0.06/s-1; and the transformation efficiency of this enzyme to 17β-estradiol was over 95.8% in 5 min. Its optimal reaction temperature was 42℃ and the optimal pH was 8.0. Divalent ions had different effect on the enzymatic activity; Mg2+ and Mn2+ could enhance the enzymatic activity.[Conclusion] The 3-oxoacyl-ACP reductase (ANI01589.1) could catalyze the transformation of 17β-estraiol efficiently and was important for the estrogen metabolism of P. putida SJTE-1.
Keywords: 3-oxoacyl-ACP reductase17β-estradiolestroneenzymatic activitytransformation efficiency
恶臭假单胞菌SJTE-1中转化17β-雌二醇的3-酰基-ACP还原酶的鉴定与功能研究
彭万里, 古丽米娜, 郑达宁, 梁如冰


微生物代谢国家重点实验室, 上海交通大学生命科学技术学院, 上海 200240
收稿日期:2018-10-26;修改日期:2018-12-19;网络出版日期:2019-03-13
基金项目:国家自然科学基金(31570099,31370152)
通信作者:梁如冰, Tel:+86-21-34204192; E-mail:icelike@sjtu.edu.cn.
摘要:[目的] 假单胞菌SJTE-1可高效转化17β-雌二醇,但是催化该转化的酶尚不清楚。本文鉴定了该菌株的一个新的3-酮酰基-ACP还原酶(ANI01589.1),并对其进行了功能研究。[方法] 首先,我们克隆了该3-酰基-ACP还原酶的编码基因,在大肠杆菌BL21(DE3)菌株中进行了异源表达;利用金属离子亲和层析法,纯化获得了重组蛋白。体外检测了重组蛋白的活性与酶学性质,并利用高效液相色谱法(HPLC)测定了该酶的催化产物。[结果] 3-酮酰基-ACP还原酶可被17β-雌二醇诱导表达,重组蛋白纯化量可达19.6 mg/L。蛋白序列比对结果表明,该蛋白包含短链脱氢酶/还原酶(SDR)的2个共有区域和多个保守残基。该酶以NAD+为辅助因子,将17β-雌二醇转化为雌酮;其Km值为0.071 mmol/L,kcat值为2.4±0.06/s-1,5 min内可转化超过95.8%的雌二醇。该酶的最佳反应温度为42℃,最佳pH为8.0。不同二价离子对该酶的活性影响不同,Mg2+和Mn2+可增强其酶活性。[结论] 这一假单胞菌SJTE-1来源的3-酮酰基-ACP还原酶可高效催化17β-雌二醇的转化,该酶可能在该菌株的雌激素代谢过程中起到重要作用。
关键词:3-酮酰基-ACP还原酶17β-雌二醇雌酮酶活转化效率
Environmental estrogens (EEs) are one of the most important environmental contaminants, causing serious environmental problem[1]. In all the natural estrogens, 17β-estradiol (E2) is with the highest estrogenic activity and influences the physiological processes of animals and humans[2]. Biodegradation is considered to be the promising strategy for the removal of EEs pollution. Microorganisms could utilize estrogens as their carbon sources or energy sources, and degrade them into non-estrogenic chemicals[3]. Strains of bacteria, fungi and algae have been found with the estrogen-degrading capabilities, and their degradation efficiencies, degradation periods and substrate spectrums varied a lot[4-8]. In most bacteria, transformation from 17β-estradiol to estrone was considered as the first and restriction step for the metabolism of 17β-estradiol. In human, hydroxysteroid dehydrogenases (HSDs), belonging to short chain dehydrogenase (SDR) family, are responsible for the mutual transformation of the active/inactive forms of estrogens through the NAD(P)H-linked oxidization/reduction[9]. In bacteria, several 3, 17β-hydroxysteroid dehydrogenases (3, 17β-HSDs) have also been found to participat in the oxidation/reduction processes of steroids[10-11]. However, because of various genetic backgrounds and complex metabolic networks in different bacteria, only few enzymes responsible for the transformation of 17β-estradiol in bacteria has been identified, and their enzymatic characteristics were still not unclear.
P. putida SJTE-1 (CGMCC NO. 6585) has been confirmed to be capable of utilizing 17β-estradiol efficiently and transforming it into the non-estrogenic chemicals. Its whole genome sequence and the proteomics analysis have been studied[7, 12]. Here we identified a new 3-oxoacyl-ACP reductase (A210_02810) of this strain induced by 17β-estradiol. Enzymatic characteristics analysis showed that this enzyme could efficiently catalyze the transformation of 17β-estradiol in vitro. These results implied that this 3-oxoacyl-ACP reductase was important for the degradation of 17β-estradiol in P. putida SJTE-1.
1 Materials and Methods 1.1 Strains, chemicals and cultures P. putida strian SJTE-1 was isolated and stored by our lab[7].E. coli strain DH5α and strain BL21 (DE3) were purchased from Invitrogen (USA) and Novagen (USA). DNA Gel Extraction kit, Plasmid Mini-prep kit and Bacterial Genome DNA extraction kit were purchased from TIANGEN (Beijing, China). T4 DNA ligase and restriction enzymes were products of TaKaRa Biocompany (Dalian, China). The Total RNA Extraction Reagents, the PrimeScript Reverse Transcriptase Kit and the Premix Ex Taq (Probe qPCR) were purchased from TaKaRa (Dalian, China). The Ni-NTA resin and BCA Protein assay kit were the product of Bio-Rad (CA, USA). Tryptone and yeast extract were from Oxoid (Basingstoke, UK). 17β-estradiol, estrone, estriol and testosterone (of HPLC grade, > 99%) were obtained from Sigma-Aldrich (St. Louis, USA). Estrogens were dissolved in dimethyl sulfoxide (DMSO) to form 5 mg/mL solutions. All other reagents were of analytical reagent grade purchased from Sigma-Aldrich (Saint Louis, USA). Luria-Bertani (LB) medium (tryptone 10.0 g/L, yeast extract 5.0 g/L, NaCl 10.0 g/L) and the minimal medium (K2HPO4 3.815 g/L, KH2PO4 0.5 g/L, (NH4)2HPO4 0.825 g/L, KNO3 1.2625 g/L, Na2SO4 0.2 g/L, CaCl2 0.02 g/L, FeCl3 0.002 g/L, MgCl2 0.02 g/L) were used for strain culture. All oligonucleotides used for gene amplification, plasmid construction, reverse transcription and quantitative PCR (RT-qPCR) were synthesized at Invitrogen (Shanghai, China).
1.2 Reverse transcription and quantitative PCR (RT-qPCR) detection The transcription profiles of the gene (A210_02810) encoding 3-oxoacyl-ACP reductase in P. putida strain SJTE-1 cultured with different carbon sources were detected with RT-qPCR. Strains were cultured in the minimal medium with glucose or E2 as its sole carbon source to the mid-exponential phase and the total RNA was extracted according to the instructions; the RNA yield was estimated using a Nanodrop UV spectrometer (Thermo Scientific, DE, USA). The reverse transcription was achieved using the PrimeScript Reverse Transcriptase kit (TaKaRa, Dalian, China) with 1 μg RNA and 20 ng random primers. The quantitative PCR was performed using the Premix Ex Taq (Probe qPCR) (TaKaRa, Dalian, China) and gene-specific primers (F: 5′-tgcaagcctgcaccggcca-3′, R: 5′-cgttgtgcacc acgatatcg-3′) in the IQTM 5 Multicolor Real-time PCR Detection System (Bio-Rad, CA, USA). The cycling conditions were 95 ℃ for 3 min, and 40 cycles of 95 ℃ for 10 s, 57 ℃ for 20 s and 72 ℃ for 20 s. A final melting analysis was obtained by slow heating with 10 s increments of 0.5 ℃ from 55 ℃ to 95 ℃. The threshold cycle (Cq) value of each sample and the relative fold change in mRNA quantity was calculated using the DDCt method[13]. Five independent experiments were performed for each RNA sample and the average values with the standard errors were calculated.
1.3 Construction of the recombinant plasmid The genome DNA of P. putida strain SJTE-1 was obtained and used for the template to amplify the encoding gene of 3-oxoacyl-ACP reductase (A210_02810, 1350 bp) with the primer pairs. The sequence of the forward primer was 5′-GGGG GGCATATGATGAGCGATCGCTACCTCGG-3′ (underlined, Nde Ⅰ site), and sequence of the reverse primer was 5′-GGGGGGGAATTCTCAAGCCCCC ATCAGTGCC-3′ (underlined, EcoR Ⅰ site). The PCR program was first denaturized at 95 ℃ for 5 min, then denaturized at 95 ℃ for 1 min, annealed at 55 ℃ for 1 min and elongated at 72 ℃ for 1.5 min; repeated this process for 30 times, and further elongated at 72 ℃ for another 5 min. The recombinant plasmid pET28a-2810 was constructed with the ligation of the restriction enzymes (EcoR Ⅰ and Nde Ⅰ) treated PCR fragments and plasmid pET28a (Novagen, USA) with T4 DNA ligase; and chemical transformation was performed to E. coli DH5α to obtain the target plasmid. All the constructs were confirmed by sequencing (Invitrogen, Shanghai, China).
1.4 Sequence alignment and structure-homology modeling Multiple sequences alignment of the 3-oxoacyl- ACP reductases and SDRs from different organisms was performed with ClustalW software. The structure-homology modeling of the 3-oxoacyl-ACP reductases was performed in SWISS-MODEL with the 3-oxoacyl-ACP reductase (4fw8.1.A) as a template.
1.5 Heterogeneous expression and affinity purification of the recombinant 3-oxoacyl-ACP reductase protein Plasmid pET28a-2810 was transformed into the competent E. coli BL21(DE3) cells. Single colony was inoculated to 5 mL LB media with 50 μg/mL kanamycin and cultured overnight at 37 ℃. The overnight cultures were transferred into 500 mL fresh LB media with 50 μg/mL kanamycin, and cultured at 37 ℃ until OD600=0.3. 0.1 mmol/L isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the culture and then induced at 37 ℃ for 3 h. Cells were harvested by centrifugation at 8000 r/min for 10 min at 4 ℃, and re-suspended with 25 mL ice-cold lysis buffer (20 mmol/L Tris-HCl, 300 mmol/L NaCl, 5 mmol/L imidazole, 5 mmol/L β-ME, 1 mmol/L PMSF, 10% glycerol, pH 8.0). The cell solution was undertaken by sonication on ice and the cell lysis was clarified by centrifugation at 12000 r/min for 30 min at 4 ℃. The cell supernatants were loaded for the affinity chromatography of Ni-NTA resin at 4 ℃ and washed with certain column of washing buffer (20 mmol/L Tris-HCl, 300 mmol/L NaCl, 5 mmol/L β-ME, 10% glycerol, 1 mmol/L PMSF, and 20–50 mmol/L imidazole, pH 8.0). Finally the recombinant protein was eluted from column using elution buffer (20 mmol/L Tris-HCl, 300 mmol/L NaCl, 5 mmol/L β-ME, 10% glycerol, 1 mmol/L PMSF, 150 mmol/L imidazole, pH 8.0). All the eluted fractions were analyzed by 15% SDS-PAGE and then stained with Coomassie Brilliant Blue R250 (Sigma-Aldrich, Saint Louis, USA). The eluted recombinant protein was stored in the storage buffer (20 mmol/L Tris-HCl, 50 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L DTT, 50% glycerol, pH 8.0) at –80 ℃. The concentration of the recombinant protein was determined using the BCA protein assay kit (TaKaRa, Dalian, China).
1.6 Enzymatic assay of the recombinant 3-oxoacyl-ACP reductase The enzymatic activity of the recombinant 3-oxoacyl-ACP reductase protein was measured by detecting the yield of co-product NADH at 355 nm (excitation)/460 nm (emission) as described previously[14]. The reaction system contained 1× reaction buffer (20 mmol/L Tris-HCl, 25 mmol/L NaCl), 200 μmol/L NAD+, different concentrations of the recombinant protein and different estrogens (17β-estradiol, estrone, estriol and testosterone). A reaction mixture without estrogen was used as blank. The parameters of 3-oxoacyl-ACP reductase (Km, Vmax, optimal pH and optimal temperature) in the transformation of 17β-estradiol were detected. The effect of different metal ions (Mg2+, Mn2+, Cu2+, Ca2+, Zn2+ and Ni2+) on the enzymatic activity were also determined. All the experiments were repeated five times and the results were the average values with standard deviations.
1.7 HPLC detection of the enzymatic reaction products The product of 17β-estradiol catalyzed by this recombinant protein was determined. The solutions of enzymatic reaction were extracted with ethyl acetate, dried in the nitrogen blowing apparatus, dissolved with acetonitrile, and detected with HPLC (High Performance Liquid Chromatography, Agilent 1260 Infinity LC, USA). The Agilent exclipse plus C18 column (3.5 μm, 4.6 mm×150.0 mm, Agilent, USA) and Diode Array Detector (DAD) detector was used with the mobile phase of acetonitrile and water (1:1, V/V) at a flow rate of 1 mL/min at 30 ℃, and the UV wavelengths were 280 nm. Five independent experiments were performed, and results were calculated as the average values with standard deviations. The quantity of steroids was calculated from their respective peak areas by using a standard curve of individual standards. The R2 values for the standard curves were > 0.99. The presented data are the averages of five independent measurements.
2 Results and Discussion 2.1 The 3-oxoacyl-ACP reductase (ANI01589.1) of P. putida SJTE-1 was induced by 17β-estradiol P. putida SJTE-1 could utilize several typical estrogens like 17β-estradiol and degrade it into non-estrogenic chemicals, whose whole genome sequence has been obtained (Genome accession number: CP015876.1)[7]. One gene (A210_02810: 622761..624113) encoding 3-oxoacyl-ACP reductase (ANI01589.1) was identified, as its transcription levels increased about 2.6 folds when P. putida SJTE-1 was cultured in minimal medium supplied with 10 μg/mL of 17β-estradiol. When estriol, estrone and testosterone were used, only little induction on the transcription of this gene was observed (Figure 1). These results implied that this 3-oxoacyl-ACP reductase of P. putida SJTE-1 induced by 17β-estradiol may participate in the utlization of this chemical.
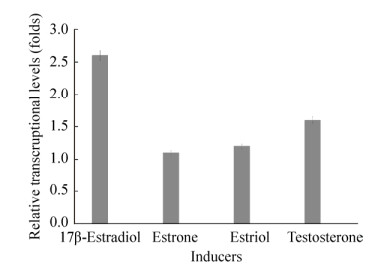 |
| Figure 1 Transcription analysis of the gene (A210_02810) encoding 3-oxoacyl-ACP reductase. The transcription level of gene A210_02810 in P. putida SJTE-1 cultured in the minimal medium with 10 μg/mL 17β-estradiol, estriol, estrone or testosterone as carbon source for 2 h were detected. The transcriptional level of this gene in P. putida SJTE-1 cultured in in the minimal medium with glucose was denoted as 1.0. |
| 图选项 |
2.2 The 3-oxoacyl-ACP reductase (ANI01589.1) was a member of SDR family This 3-oxoacyl-ACP reductase (ANI01589.1) of P. putida SJTE-1 contained 450 amino acids. Multiple sequences alignment showed that this protein contained two conserved motifs identical to the members of the SDR superfamily, the co-factor binding Rossmann-fold motif (Gly-X-X-X-Gly- X-Gly) and the active Tyr-X-X-X-Lys (YXXX) motif. The critical catalytic Tyr residue (residues 356) in YXXXK active site motif served as a proton acceptor, and the upstream Ser (residues 343) and Asn (residues 315) contributed to the active site (Figure 2-A)[15]. Its secondary structure was composed of the βαβ units like a parallel β-sheet sandwiched between two α-helixes, consistent with those of other SDRs from E. coli, C. testosteroni, Rhodococcus sp., and Homo sapiens (Figure 2-A)[16]. Structure modelling results indicated that this 3-oxoacyl-ACP reductase showed 52.05% identity to its template, 3-oxoacyl-(Acyl-carrier-protein) reductase (4fw8.1.A). Its co-factor binding motif located in the central and its C-terminal located on the surface (Figure 2-B). Therefore, this 3-oxoacyl-ACP reductase was a member of SDR superfamily.
 |
| Figure 2 Multiple sequences alignment and structure modelling of the 3-oxoacyl-ACP reductase. A: The 3-oxoacyl-ACP reductase from P. putida SJTE-1 (ANI01589.1), Rhodococcus sp. P14 (WP_010595922.1), E. coli MG1655 (WP_001499617.1), Homo sapiens (XP_005263372.1), C. testosteroni ATCC11996 (WP_003080542.1) and P. aeruginosa PAO1 (NP_250519.1) were aligned with ClustalW. The secondary structure of seven α-helixes and seven β-strands was marked. B: The three-dimension structure modelling result of this 3-oxoacyl-ACP reductase (ANI01589.1), performed with SWISS-MODEL with the 3-oxoacyl-ACP reductase (4fw8.1.A) as template. |
| 图选项 |
2.3 The recombinant 3-oxoacyl-ACP reductase could transform 17β-estradiol efficiently The recombinant 3-oxoacyl-ACP reductase was purified by the Ni-NTA affinity chromatography. The purified protein was detected as a single band with the apparent molecular mass of about 49.5 kDa and the purity was no less than 98% (Figure 3-A). More than 19.6 mg of the recombinant 3-oxoacyl-ACP reductase was obtained from 1 liter cell culture. The specific activity of the purified protein in the transformation of 17β-estradiol was 18.23 U/mg.
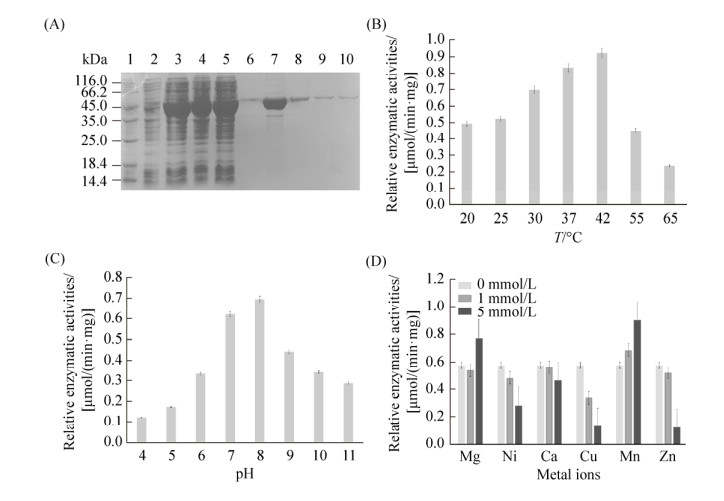 |
| Figure 3 The purification and characterization of 3-oxoacyl-ACP reductase in vitro. A: SDS-PAGE detection of recombinant 3-oxoacyl-ACP reductase. Lane 1: protein marker; lane 2: cell lysis solution before induction; lane 3: cell lysis solution after induction; lane 4–5: the solution after column loading; lane 6–10: the eluted protein solution. B: The relative initial rates of this enzyme at different temperatures were detected with E2 as substrate. C: The relative initial rates of this enzyme under different pH were detected with E2 as substrate. D: The relative initial rates of this enzyme with different metal ions were detected with E2 as substrate. |
| 图选项 |
The kinetic parameters for the recombinant 3-oxoacyl-ACP reductase were measured by detecting the fluorescence variation of NAD(P)H at 460 nm. Results showed that NAD+ not NADP+ was the cofactor of this recombinant protein, and only 17β-estradiol and testosterone could be oxidized by this enzyme when different estrogens (17β-estradiol, estriol, estrone and testosterone) were used. The production of NADH with 17β-estradiol as substrate were more efficient than that with testosterone. These results implied that the prefered substrate of this recombinant 3-oxoacyl-ACP reductase was 17β-estradiol (Table 1).
Table 1. The substrates spectrum of the recombinant 3-oxoacyl-ACP reductase
| Substrate | 17β-estradiol | Testosterone | Estriol | Estrone |
| Fluorescence of NADH | 0.984 | 0.430 | 0.025 | 0.041 |
表选项
Then the Km value and Vmax value of this recombinant protein for E2 transforamtion were determined. The Km value of this enzyme for E2 oxidation was 0.071±0.020 mmol/L and its Vmax value was 0.72±0.03 μmol/(s·mg); its kcat value was 2.4±0.06/s and the specificity constant (kcat/Km) for NAD+ was 33.80 mmol/(s·L). However, the Km value of this enzyme for for E1 reduction was 0.79±0.06 mmol/L and its Vmax value was 0.11±0.05 μmol/(s·m) (Table 2). These results meant that this recombinant 3-oxoacyl-ACP reductase was mainly responsible for the oxidation reaction of 17β-estradiol, other than the reduction of estrone.
Table 2. The enzymatic characteristics of the recombinant 3-oxoacyl-ACP reductase
| Reactions | Km/(mmol/L) | Vmax/[(μmol?mg)/s] | Kcat/s–1 | Kcat/Km/[L/(s?mmol)] |
| Oxidation with E2 as substrate, with NAD+ as cofactor | 0.071± 0.020 | 0.72±0.03 | 2.40±0.06 | 33.80 |
| Reduction with E1 as substrate, with NADH as cofactor | 0.790±0.060 | 0.11±0.05 | 0.75±0.03 | 0.95 |
表选项
Enzymatic characteristics analysis demonstrated that the optimal reaction temperature of this recombinant protein was 42 ℃ and the optimal reaction pH was 8.0 (Figure 3-B, 3-C). The effect of different divalent ions (Mg2+, Mn2+, Ca2+, Zn2+, Cu2+ and Ni2+) on the enzymatic activity were also analyzed. Mg2+ and Mn2+ could stimulate the oxidation activity of this enzyme for E2 a little, while other ions of low concentration could inhibit the enzymatic activity significantly (Figure 3-D). Further HPLC detection showed that estrone was the product of the oxidation reaction when E2 used as substrate, and the transformation efficiency was over 95.8% of E2 (0.2 μmol/L) in five minutes (Figure 4). Therefore, this 3-oxoacyl-ACP reductase could oxidize 17β-estradiol into estrone efficiently, implyinging its involvement in the estrogen metabolism of P. putida SJTE-1.
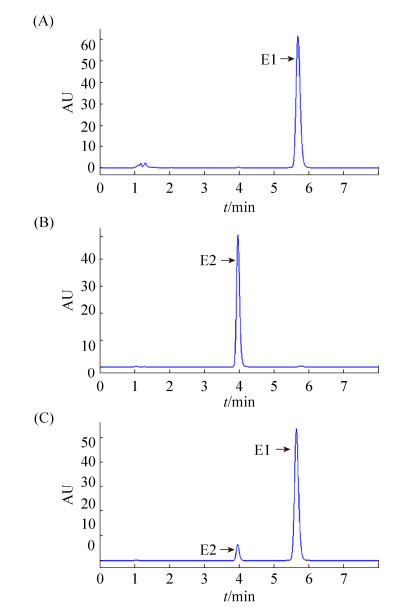 |
| Figure 4 HPLC results of the reaction product catalyzed by the recombinant 3-oxoacyl-ACP reductase. A: The HPLC trace of 0.2 μmol/L estrone (E1); B: The HPLC trace of 0.2 μmol/L 17β-estradiol (E2); C: The HPLC trace of the 3-oxoacyl-ACP reductase oxidation reaction with 17β-estradiol as substrate. The reaction mixture contained 0.2 μmol/L 17β-estradiol, 0.4 μmol/LNAD+, and 0.3 μmol/L enzyme in 1×reaction buffer (20 mmol/L Tris-HCl, 25 mmol/L NaCl), and the reaction was performed at 37 ℃ for 2.5 min. |
| 图选项 |
Environmental estrogens pollution was considered as one of the great environment problem, and biodegadation with microorganisms was used for efficient environmental remediation. In these bacteria, the HSDs belonging to SDR family were considered as the first restriction enzyme to start the transformation of steroids[10-11]. In this work, the identified 3-oxoacyl-ACP reductase of P. putida SJTE-1 could efficiently catalyze the transformation of 17β-estradiol, and its enzymatic activity was similar to or higher than those of reported HSDs[10-11]. As it was induced by 17β-estradiol, this 3-oxoacyl-ACP reductase was probably involved in the transformation of this chemical in P. putida SJTE-1.
Proteins of SDR family share some conserved structures although their identity was low[16]. Besides the classic motifs GXXXGXG and YXXXK, the classical SDRs also contained four highly conserved residues (Asn-Ser-Tyr-Lys) to form the catalytic center; and NAD(P)/NAD(P)H always acted as their reaction co-factors[16]. A typical structure, seven parallel β-sheets of the center and three α-helixes on each side, was conserved in the HSD of SDR family. As to the 3-oxoacyl-ACP reductase of P. putida SJTE-1 studied in this work, the conserved motifs, the βαβ units of SDR family and the conserved Ser-Tyr-Lys-triad of SDRs were all observed in its sequence and structure; thus, it was a typical SDR.
Although several 3, 17β-HSDs have been reported to participate in the metabolism of steroids, the catalyzing enzyme for 17β-estradiol transformation was still not very clear. The reason was that most of the identified 3, 17β-HSDs could not be induced by 17β-estradiol, although in vitro catalyzation was observed[11]. Multiple isoenzymes may participate in the transformation of 17β-estradiol in vivo. The 3-oxoacyl-ACP reductase (EC1.1.1.100) is known as a component of fatty acid synthetase. Two isoforms, NADPH-dependence and NADH-dependence, have been reported; the NADPH-dependence enzyme is confirmed to be involved in fatty acid biosynthesis, while the function of the NADH-dependence enzyme is unclear[17-18]. Previously a 3-oxoacyl-ACP reductase from C. testosteroni ATCC11996 has been proved to be important for the degradation of testosterone; however, it could not be induced by estradiol[19]. The 3-oxoacyl-ACP reductase identified in this work could be efficiently induced by 17β-estradiol, probably important for estrogen degradation in P. putida SJTE-1. Considering its high enzymatic activity, the transformation of 17β-estradiol could be finished efficiently in P. putida SJTE-1.
References
| [1] | Combalbert S, Hernandez-Raquet G. Occurrence, fate, and biodegradation of estrogens in sewage and manure. Applied Microbiology and Biotechnology, 2010, 86(6): 1671-1692. DOI:10.1007/s00253-010-2547-x |
| [2] | Luine VN. Estradiol and cognitive function: past, present and future. Hormones and Behavior, 2014, 66(4): 602-618. DOI:10.1016/j.yhbeh.2014.08.011 |
| [3] | Yu CP, Deeb RA, Chu KH. Microbial degradation of steroidal estrogens. Chemosphere, 2013, 91(9): 1225-1235. DOI:10.1016/j.chemosphere.2013.01.112 |
| [4] | Haiyan R, Shulan J, ud din Ahmad N, Wang D, Cui CW. Degradation characteristics and metabolic pathway of 17α-ethynylestradiol by Sphingobacterium sp. JCR5. Chemosphere, 2007, 66(2): 340-346. DOI:10.1016/j.chemosphere.2006.04.064 |
| [5] | Hom-Diaz A, Llorca M, Rodríguez-Mozaz S, Vicent T, Barceló D, Blánquez P. Microalgae cultivation on wastewater digestate: β-estradiol and 17α-ethynylestradiol degradation and transformation products identification. Journal of Environmental Management, 2015, 155: 106-113. |
| [6] | Shi JH, Fujisawa S, Nakai S, Hosomi M. Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea. Water Research, 2004, 38(9): 2323-2330. DOI:10.1016/j.watres.2004.02.022 |
| [7] | Liang RB, Liu H, Tao F, Liu Y, Ma C, Liu XP, Liu JH. Genome sequence of Pseudomonas putida strain SJTE-1, a bacterium capable of degrading estrogens and persistent organic pollutants. Journal of Bacteriology, 2012, 194(17): 4781-4782. DOI:10.1128/JB.01060-12 |
| [8] | Yang J, Li WJ, Ng TB, Deng XZ, Lin J, Ye XY. Laccases: production, expression regulation, and applications in pharmaceutical biodegradation. Frontier in Microbiology, 2017, 8: 832. DOI:10.3389/fmicb.2017.00832 |
| [9] | Chang YH, Wang YL, Lin JY, Chuang LY, Hwang CC. Expression, purification, and characterization of a human recombinant 17β-hydroxysteroid dehydrogenase type 1 in Escherichia coli. Molecular Biotechnology, 2010, 44(2): 133-139. DOI:10.1007/s12033-009-9221-5 |
| [10] | Ye XY, Wang H, Kan J, Li J, Huang TW, Xiong GM, Hu Z. A novel 17β-hydroxysteroid dehydrogenase in Rhodococcus sp. P14 for transforming 17β-estradiol to estrone. Chemico-Biological Interactions, 2017, 276: 105-112. DOI:10.1016/j.cbi.2017.06.010 |
| [11] | Yu YH, Liu CZ, Wang BX, Li YH, Zhang H. Characterization of 3, 17β-hydroxysteroid dehydrogenase in Comamonas testosteroni. Chemico-Biological Interactions, 2015, 234: 221-228. DOI:10.1016/j.cbi.2015.01.005 |
| [12] | Xu J, Zhang L, Hou JL, Wang XL, Liu H, Zheng DN, Liang RB. iTRAQ-based quantitative proteomic analysis of the global response to 17β-estradiol in estrogen-degradation strain Pseudomonas putida SJTE-1. Scientific Reports, 2017, 7: 41682. DOI:10.1038/srep41682 |
| [13] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 2001, 25: 402-408. DOI:10.1006/meth.2001.1262 |
| [14] | Pan TY, Huang P, Xiong GM, Maser E. Isolation and identification of a repressor TetR for 3, 17β-HSD expressional regulation in Comamonas testosteroni. Chemico-Biological Interactions, 2015, 234: 205-212. DOI:10.1016/j.cbi.2014.12.034 |
| [15] | Beck KR, Kaserer T, Schuster D, Odermatt A. Virtual screening applications in short-chain dehydrogenase/reductase research. Journal of Steroid Biochemistry and Molecular Biology, 2017, 171: 157-177. DOI:10.1016/j.jsbmb.2017.03.008 |
| [16] | Kavanagh KL, J?rnvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and Molecular Life Sciences, 2008, 65(24): 3895-3906. DOI:10.1007/s00018-008-8588-y |
| [17] | Safford R, Windust JHC, Lucas C, De Silva J, James CM, Hellyer A, Smith CG, Slabas AR, Hughes SG. Plastid-localised seed acyl-carrier protein of Brassica napus is encoded by a distinct, nuclear multigene family. FEBS Journal, 1988, 174(2): 287-295. |
| [18] | Slabas AR, Chase D, Nishida I, Murata N, Sidebottom C, Safford R, Sheldon PS, Kekwick RG, Hardie DG, Mackintosh RW. Molecular cloning of higher-plant 3-oxoacyl-(acyl carrier protein) reductase. Sequence identities with the nodG-gene product of the nitrogen-fixing soil bacterium Rhizobium meliloti. Biochemical Journal, 1992, 283: 321-326. DOI:10.1042/bj2830321 |
| [19] | Zhang H, Ji Y, Wang Y, Zhang X, Yu YH. Cloning and characterization of a novel β-ketoacyl-ACP reductase from Comamonas testosteroni. Chemico-Biological Interactions, 2015, 234: 213-220. DOI:10.1016/j.cbi.2015.01.003 |
