原野, 胡彦波, 周义发


东北师范大学生命科学学院, 吉林 长春 130024
收稿日期:2017-03-31;修回日期:2017-05-27;网络出版日期:2017-06-27
基金项目:吉林省自然科学基金(20160101343JC)
*通信作者:周义发, Tel/Fax:+86-431-85098212;E-mail:zhouyf383@nenu.edu.cn
摘要:糖苷广泛存在于自然界,具有多种药理活性,是人类发现与生产药物的重要来源。糖苷中糖链部分的组成与其药理活性密切相关,改变糖苷分子中的糖链结构能改变糖苷的药理活性,为开发药物提供更多的化合物资源。糖苷水解酶修饰糖链具有效率高、成本低、污染小等优点,被广泛应用于活性糖苷与苷元的制备。本文系统地总结了糖苷水解酶转化制备活性糖苷与苷元的研究进展,为研究糖苷酶生物转化制备活性化合物提供数据资源,为相关的研究和生产提供有用的文献资料。
关键词: 糖苷 糖苷水解酶 生物转化
Glycosidase:An effective tool for the preparation of active glycosides and aglycone
Ye Yuan, Yanbo Hu, Yifa Zhou


School of Life Sciences, Northeast Normal University, Changchun 130024, Jilin Province, China
Received 31 March 2017; Revised 27 May 2017; Published online 27 June 2017
*Corresponding author: Yifa Zhou, Tel/Fax:+86-431-85098212;E-mail:zhouyf383@nenu.edu.cn
Supported by the Natural Science Foundation of Jilin Province (20160101343JC)
Abstract: Glycosides widely distributed in nature have many pharmacological activities. A lot of drugs belong to glycoside and the sugar chains are closely related to its pharmacological activities. The modification of sugar chains can change the pharmacological activities of glycosides, which provides tremendous glycoside resource for drug development. Because of high efficiency and low pollution, biotransformation by glycosidase is widely used in the preparation of active glycosides. This review summarized recent advances in the preparation of active glycosides by glycosidase transformation, and provided useful references for preparing active glycosides and related research work.
Key words: glycosides glycosidase biotransformation
糖苷是糖或糖衍生物的半缩醛羟基与另一非糖物质缩合而成的化合物。天然糖苷主要来自植物的次生代谢产物。出于自身的防御需要,植物合成大量的糖苷[1],为人类提供了研究、开发和生产药物候选化合物的丰富资源。糖苷类化合物具有祛风湿、抑菌、抗炎、抗肿瘤、免疫调节、改善呼吸道及消化道等多种重要的药理活性[2-4],目前市场销售的药物约有70%以上与糖苷类化合物相关。随着糖工程的兴起[5],以及分离纯化和鉴定技术的发展,对于植物中糖苷类有效成分的研究将更为深入,应用将更加广泛。根据结构不同,糖苷有多种分类方式。按苷原子的不同可分为氧糖苷、硫糖苷、碳糖苷和氮糖苷,其中氧糖苷最为常见。糖苷结构多样性产生多种药理活性[6]。糖苷的药理活性不仅与苷元有关,与糖链部分也密切相关。糖苷中糖链的单糖组成、糖苷键构型、糖基连接方式等都影响糖苷的活性及代谢途径[7],有些糖苷通过水解生成苷元,产生更好的药理活性,如槲皮素、京尼平等。修饰糖苷分子中的糖链部分,研究其构效关系,对发现新型的糖苷药物具有重要意义。利用酸或碱催化等化学法能够水解糖链,也可以采用微生物以及酶催化等生物学方法水解糖链。化学法水解糖苷有时会产生较多的副产物,同时容易引起环境污染,而生物学方法可能克服这些问题。因此,糖苷水解酶被认为是制备活性糖苷与苷元潜在的有效工具。本文系统地总结了糖苷水解酶转化制备活性糖苷与苷元的研究进展。
1 糖苷水解酶概述 糖苷水解酶是真正意义上的水解酶,不需要任何辅酶和辅因子[8]。糖苷水解酶数量众多,广泛存在于细菌、真菌、植物种子和动物器官中,根据氨基酸残基序列和结构的相似性可划分为不同家族,目前已有145个糖苷水解酶家族被报道[9]。
糖苷水解酶的性质和功能一直是糖生物学领域研究的热点。我国对糖苷水解酶的研究始于20世纪50年代末,张树政院士等分析比较了酒精工业中不同曲霉的淀粉酶系的组成,在国内首先用纸电泳法分离测定了淀粉酶。1966年从150株根霉中筛选出3株产高活力淀粉葡萄糖苷水解酶的根霉,并对其酶活性质进行初步探究[10]。80年代开始,张树政院士开展了多种糖苷酶的基础和应用研究,大力倡导糖生物学和糖工程前沿计划。作为糖生物学的奠基人之一,张树政院士长期致力于微生物生物化学和糖生物学的研究,在糖苷酶的结构与功能、糖生物学和糖生物工程学中取得了显著的成就,为我国的酶制剂工业及酶学的发展做出了奠基性的贡献。
随着糖类化合物在生物学领域的重要性越发凸显,有关糖苷水解酶的研究与应用也越来越受到关注。目前,糖苷水解酶主要是通过分离纯化和分子克隆技术从微生物或动植物体中获得的。于巍等[11]从土壤微生物中筛选得到Enterobactercloacae YW2112菌株,从中分离纯化的糖苷酶能特异性水解神经节苷脂中连接神经酰胺和寡糖链之间的糖苷键,是研究神经节苷脂结构与功能的重要工具。张树政等[12]从巨大芽孢杆菌的全基因组DNA文库中重组构建了β-淀粉酶,经氨基酸序列比较分析发现该酶依次由信号肽域、糖基水解酶催化功能域和淀粉结合域3个功能结构域组成。随着分子生物学技术的不断发展,随机突变和定向进化在改良糖苷水解酶中应用广泛。中国科学院微生物研究所唐双焱等[13]通过DNA重组技术提高了芽孢杆菌糖化酶的热稳定性,并预测出突变酶热稳定性提高的机理。目前,糖苷水解酶的制备和工业化应用已取得显著成效。糖苷水解酶易通过发酵生产获得,随着基因工程和蛋白质工程的快速发展,重组糖苷水解酶由于具有表达量高、易纯化等特点被广泛使用。酶法转化过程条件温和、专一性好、产量高、绿色环保,因此糖苷水解酶成为转化制备活性糖苷与苷元的有效工具。
2 氧糖苷的生物转化 目前,糖苷水解酶被应用于多种活性糖苷与苷元制备的研究。其中,氧糖苷中的皂苷和黄酮苷的酶法制备研究最为广泛。经过科研工作者们多年的努力,糖苷水解酶制备活性糖苷与苷元取得了诸多可喜的成果[14-16]。
2.1 皂苷的生物转化 皂苷是苷元为三萜或甾烷类化合物的一类糖苷,是人参、甘草和薯蓣等许多中草药的有效成分之一(主要皂苷结构见图 1),具有抗氧化、抗肿瘤、抗衰老、增强免疫力等多种功效。文献中对人参皂苷生物转化研究报道较多。目前,分离鉴定出的150多种人参皂苷,在人参中含量不同,人参皂苷Rb1、Rb2、Rc、Rd、Re和Rg1等含量高达80%,而人参皂苷Rg3、Rh2、F2和Compound K (C-K)等稀有皂苷含量很少甚至没有[17]。研究表明,有些稀有皂苷具有较好的药理学活性[18]。但由于含量低,制备和生产受到限制。同一类型的人参皂苷具有相同的苷元,只是糖链不同。稀有人参皂苷与含量较高的同种类型的皂苷往往只差别2-3个糖基。因此,可以酶催化水解高含量皂苷转化制备相同类型的活性稀有皂苷。
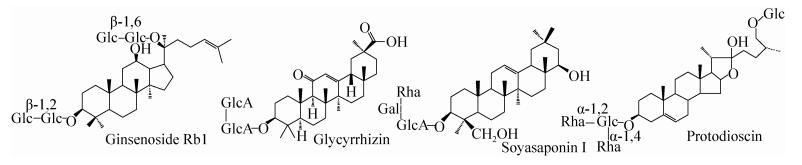 |
| 图 1 主要皂苷结构示意图 Figure 1 Structure of main saponins. |
| 图选项 |
不同的糖苷水解酶具有不同的选择性,水解人参皂苷的路径也不相同。如表 1所示,利用不同的糖苷水解酶可以制备不同的稀有人参皂苷。通过水解人参皂苷Rb1、Rb2、Rb3和Rc的C-20位外侧糖基可以制备人参皂苷Rd。从中国白玉蜗牛(China white jade snail)[19]和Thermus caldophilus[20]中分离纯化的β-葡萄糖苷水解酶能够实现人参皂苷Rb1到Rd的转化,Kim等[21]利用分子克隆技术从土壤微生物中获得能够将人参皂苷Rb1转化为Rd的重组糖苷水解酶。随后,科研工作者从Thermotoga thermarum[22]和Bifidobacterium longum H-1[23]中克隆得到葡萄糖苷酶,提高了转化制备人参皂苷Rd的效率。利用重组技术从Flavobacteriumjohnsoniae[24]和Thermus thermophilus[25]中获得的葡萄糖苷酶不仅可以将人参皂苷Rb1转化成Rd,同样可以水解绞股蓝皂苷ⅩⅦ (G17) 的C-20位糖链,生成人参皂苷F2。除了葡萄糖苷酶外,从人参根[26]和Leuconostoc sp.[27]中获得了能转化人参皂苷Rc为Rd的α-L-阿拉伯呋喃糖苷水解酶。从Bifidobacterium breve[28]和Bifidobacterium longum[29]中得到α-L-阿拉伯呋喃糖苷水解酶和α-L-阿拉伯吡喃糖苷水解酶,能够转化人参皂苷Rc和Rb2成为Rd。有文献报道,Caldicellulosiruptor saccharolyticus[30]和Rhodanobacter ginsenosidimutans[31]中的α-L-阿拉伯呋喃糖苷水解酶不仅可以水解人参皂苷Rc为Rd,同时可以将Compound Mc1 (C-Mc1) 转化成F2。Yu等[32]从Aspergillus中分离纯化的糖苷水解酶则可将人参皂苷Rb1、Rb2、Rb3和Rc全部转化为Rd。一些糖苷水解酶能够完全水解二醇型人参皂苷Rb1、Rb2、Rb3、Rc和Rd等分子中C-20位的糖链,生成人参皂苷Rg3,使得Rg3能够大规模生产,被开发成为抗肿瘤的药物。Paecilomycesbainier[33]和Microbacterium esteraromaticum[34]中的葡萄糖苷酶能直接将人参皂苷Rb1水解成Rg3,而从Microbacterium esteraromaticum[35]中分离纯化的葡萄糖苷酶能将人参皂苷Rb2水解成Rg3。利用分子克隆技术从Pseudonocardia[36]中克隆得到的重组糖苷水解酶可转化人参皂苷Rb1、Rb3和Rd制备Rg3。同样,水解人参皂苷中C-3位的糖基也可制备一系列活性稀有人参皂苷。从Sphingomonas[37]和Sphingopyxis alaskensis[38]中克隆得到的重组葡萄糖苷酶能水解人参皂苷Rb1、Rb2、Rc、Rd和Rg3分子中C-3位糖链外侧的葡萄糖,相应地制备G17、Compound O (C-O)、C-Mc1、F2和Rh2。有些糖苷酶则能直接水解C-3位内侧葡萄糖基,如来源于Terrabacter ginsenosidimutans[39]和Esteya vermicola[40]的葡萄糖苷酶能水解人参皂苷Rb1、Rb2、Rb3、Rc和Rd分子C-3位的糖链,生成相应的皂苷LXXV (G75)、Compound Y (C-Y)、Compound Mx (C-Mx)、Compound Mc (C-Mc)和C-K。此外,一些糖苷水解酶能够同时水解二醇型人参皂苷中C-20和C-3位糖基。从Arthrobacterchlorophenolicus[41]中克隆得到的重组葡萄糖苷酶能将人参皂苷Rb1、Rb2和Rc转化成F2。Fusobacterium K60[42]、endophytic fungi GE 17-18[43]、Sulfolobusacidocaldarius[44]、Aspergillus niger[45]和Microbacteriuesteraromaticum[46]中的糖苷水解酶则能够水解人参皂苷Rb1制备C-K。
表 1. 糖苷水解酶转化人参皂苷 Table 1. Biotransformation of ginsenosides by glycosidase
| Product | Substrate | Reaction | Organism | Reference |
| Rd | Rb1 | β-Glucosidase | China white jade snail | Luan[19] |
| Rd | Rb1 | β-Glucosidase | Thermus caldophilus | Son[20] |
| Rd | Rb1 | β-Glucosidase | Uncultured bacteria | Kim[21] |
| Rd | Rb1 | β-Glucosidase | Thermotoga thermarum | Zhao[22] |
| Rd | Rb1 | β-Glucosidase | Bifidobacterium longum H-1 | Jung[23] |
| Rd | Rb1 | β-Glucosidase | Flavobacterium johnsoniae | Hong[24] |
| Rd | Rb1 | β-Glucosidase | Thermus thermophilus | Shin[25] |
| Rd | Rb1 | β-Glucosidase | Penicillium oxalicum | Gao[47] |
| Rd | Rb1 | β-Glucosidase | Cladosporium fulvum | Gao[48] |
| Rd | Rc | α-L-Arabinofuranosidase | Panax ginseng | Zhang[26] |
| Rd | Rc | α-L-Arabinofuranosidase | Leuconostoc | Liu[27] |
| Rd | Rc | α-L-Arabinofuranosidase | Bifidobacterium breve | Shin[28] |
| Rd | Rc | α-L-Arabinofuranosidase | Bifidobacterium longum | Lee[29] |
| Rd | Rc | α-L-Arabinofuranosidase | Caldicellulosiruptor saccharolyticus | Shin[30] |
| Rd | Rc | α-L-Arabinofuranosidase | Rhodanobacter ginsenosidimutans | An[31] |
| Rd | Rb2 | α-L-Arabinopyranosidase | Bifidobacterium breve | Shin[28] |
| Rd | Rb2 | α-L-Arabinopyranosidase | Bifidobacterium longum | Lee[29] |
| Rd | Rb1/Rb2/Rb3/Rc | Glycosidase | Aspergillus | Yu[32] |
| Rg3 | Rb1 | β-Glucosidase | Paecilomyces bainier | Yan[33] |
| Rg3 | Rb1 | β-Glucosidase | Microbacterium esteraromaticum | Quan[34] |
| Rg3 | Rb2 | β-Glucosidase | Microbacterium esteraromaticum | Quan[35] |
| Rg3 | Rb1/Rb3/Rd | β-Glucosidase | Pseudonocardia | Du[36] |
| G17 | Rb1 | β-Glucosidase | Sphingomonas | Wang[37] |
| G17 | Rb1 | β-Glucosidase | Sphingopyxis alaskensis | Shin[38] |
| G17 | Rb1 | β-Glucosidase | Cellulosimicrobium cellulans | Yuan[49] |
| G75 | Rb1 | β-Glucosidase | Terrabacter ginsenosidimutans | An[39] |
| G75 | Rb1 | β-Glucosidase | Esteya vermicola | Hou[40] |
| F2 | G17 | β-Glucosidase | Flavobacterium johnsoniae | Hong[24] |
| F2 | G17 | β-Glucosidase | Thermus thermophilus | Shin[25] |
| F2 | C-Mq | α-L-Arabinofuranosidase | Caldicellulosiruptor saccharolyticus | Shin[30] |
| F2 | C-Mc1 | α-L-Arabinofuranosidase | Rhodanobacter ginsenosidimutans | An[31] |
| F2 | Rd | β-Glucosidase | Cellulosimicrobium cellulans | Yuan[49] |
| F2 | Rb1/Rb2/Rc | β-Glucosidase | Arthrobacter chlorophenolicus | Park[41] |
| Rh2 | Rg3 | β-Glucosidase | Sphingopyxis alaskensis | Shin[38] |
| CK | Rd | β-Glucosidase | Terrabacter ginsenosidimutans | An[39] |
| CK | Rd | β-Glucosidase | Esteya vermicola | Hou[40] |
| CK | Rb1 | β-Glucosidase | Fusobacterium K-60 | Park[42] |
| CK | Rb1 | β-Glucosidase | endophytic fungi GE 17-18 | Fu[43] |
| CK | Rb1/Rb2 | β-Glucosidase | Sulfolobus acidocaldarius | Noh[44] |
| CK | Rb1/Rb2/Rb3/Rc | β-Glucosidase | Aspergillus niger | Liu[45] |
| CK | Rb1/Rb2 | β-Glucosidase | Microbacteriu esteraromaticum | Quan[46] |
| C-O | Rb2 | β-Glucosidase | Cellulosimicrobium cellulans | Yuan[49] |
| C-Y | Rb2 | β-Glucosidase | Terrabacter ginsenosidimutans | An[39] |
| C-Mc | Rc | β-Glucosidase | Terrabacter ginsenosidimutans | An[39] |
| C-Mci | Rc | β-Glucosidase | Cellulosimicrobium cellulans | Yuan[49] |
| C-Mx | Rb3 | β-Glucosidase | Terrabacter ginsenosidimutans | An[39] |
| Rg2 | Re | β-Glucosidase | Microbacterium esteraromaticum | Quan[50] |
| Rg2 | Re | β-Glucosidase | Mucilaginibacter | Cui[51] |
| Rg2 | Re | β-Glucosidase | Pseudonocardia | Du[36] |
| Rhi | Rgi | β-Glucosidase | Microbacterium esteraromaticum | Quan[50] |
| Rhi | Rf | β-Glucosidase | Pyrococcus furiosus | Oh[52] |
| Rhi | Rf | β-Glucosidase | Aspergillus niger | Ruan [53] |
| Rhi | Rg2 | α-L-Rhamno sidase | Absidia | Yu[54] |
| Rhi | R2 | β-Xylosidase | Thermoanaerobacterium | Shin[55] |
| Fi | Rgi | β-Glucosidase | Fusarium moniliforme | Kim[56] |
| Fi | Rgi | β-Glucosidase | Penicillium sclerotiorum | Wei[57] |
| Fi | Rgi | β-Glucosidase | Sanguibacter keddieii | Kim[58] |
| G17: gypenoside XVII; G75: gypenoside LXXV; C-O: compound O; C-Y: compound Y; C-Mc1: compound Mc1; C-Mc: compound Mc; C-Mx: compound Mx; C-K: compound K. | ||||
表选项
三醇型人参皂苷中C-6和C-20位的糖基也能被糖苷水解酶水解。人参皂苷Rg2可通过糖苷酶水解Re分子中的C-20的葡萄糖获得,利用重组技术从Microbacterium esteraromaticum[50]、Mucilaginibacter[51]和Pseudonocardia[36]中克隆得到的重组葡萄糖苷酶不仅可以将人参皂苷Re转化成Rg2,也可以将人参皂苷Rg1转化成Rh1。通过水解人参皂苷Rf、Rg2和R2的C-6位外侧的葡萄糖、鼠李糖和木糖均可以转化制备Rh1[52-55]。与人参皂苷Rh1不同,人参皂苷F1只有一个葡萄糖连在其苷元的C-20 位。Fusarium moniliforme[56]、Penicillium sclerotiorum[57]和Sanguibacter keddieii[58]中的葡萄糖苷酶可以特异性水解人参皂苷Rg1的C-6位葡萄糖,生成人参皂苷F1。
本实验室旨在研究长白山特色天然药物的有效成分及其化学性质和生物学活性,近年来,在人参皂苷生物转化研究中取得了一些进展。在真菌研究中,对植物病原真菌转化人参皂苷的效率和转化产物进行了系统分析[59],从Penicilliumoxalicum[47]和Cladosporium fulvum[48]中分离纯化出转化人参皂苷Rb1为Rd的糖苷水解酶。对于细菌的研究,本实验室从长白山林地的人参种植土壤中筛选出24种能够转化人参皂苷的细菌,并对它们的转化能力和转化产物进行了系统分析,发现Cellulosimicrobium cellulans sp. 21水解能力最强[60]。从中克隆获得的重组糖苷水解酶CcBgl1A能够特异性水解人参二醇型皂苷C-3位外侧连接的葡萄糖,将人参皂苷Rb1、Rb2、Rc和Rd完全水解为G17、C-O、C-Mc1和F2[49]。目前,我们通过组合使用多种重组糖苷水解酶实现了百克级规模制备Rg3、Rh2、C-K、Rg2和F1等多种稀有人参皂苷,为人参的开发利用和工业生产奠定了基础。
糖苷水解酶不仅应用于转化制备活性稀有人参皂苷,同时也被广泛应用于水解修饰甘草、大豆和薯蓣等皂苷(表 2)。从Streptococcus LJ-22[61]和Penicillium purpurogenum Li-3[62]中分离纯化的葡萄糖醛酸苷酶能够水解甘草皂苷生成单葡萄糖醛酸甘草酸,且没有副产物甘草次酸的生成。Morana等[63]使用来源于Aspergillus niger的葡萄糖醛酸苷酶,可将甘草皂苷彻底水解生成甘草次酸。从Aspergillus oryzae[64]中分离纯化的大豆皂苷水解酶能够水解大豆皂苷Ⅰ生成大豆皂醇B。而Neocosmospora vasinfecta[65]中的一种新大豆皂苷水解酶能将大豆皂苷Ⅰ、Ⅱ、Ⅲ都转化成大豆皂醇B,为制备具有抗氧化和调血脂的大豆皂苷提供了有效的工具。在甾体皂苷中,对薯蓣皂苷糖链水解修饰的研究比较系统。Inoue等[66]从Costusspeciosus中分离纯化出能够水解原薯蓣皂苷生成薯蓣皂苷的葡萄糖苷酶。Liu等[67]从Aspergillusoryzae中分离纯化并克隆得到重组原薯蓣皂苷水解酶,该酶能水解原薯蓣皂苷中葡萄糖基和α-1, 4鼠李糖基,生成薯蓣次苷Ⅲ。Feng等[68]从Curvularia lunata中分离纯化的α-L-鼠李糖苷酶能够水解薯蓣皂苷中的α-1, 2鼠李糖基,生成薯蓣次苷Ⅴ。Qian等[69]从新鲜牛肝中分离纯化出一种α-L-鼠李糖苷酶,该酶能够水解薯蓣皂苷中的α-1, 2和α-1, 4两个鼠李糖基,生成葡萄糖基-薯蓣皂苷元。Fu等[70]则从Absidia中分离纯化出能将薯蓣皂苷彻底水解为薯蓣皂苷元的薯蓣皂苷水解酶。
表 2. 糖苷水解酶转化其他皂苷 Table 2. Biotransformation of other saponins by glycosidase
| Product | Substrate | Reaction | Organism | Reference |
| GAMG | Glycyrrhizin | β-Glucuronidase | Streptococcus | Park[61] |
| GAMG | Glycyrrhizin | β-Glucuronidase | Penicillium purpurogenum | Zou[62] |
| Glycyrrhetinic acid | Glycyrrhizin | β-Glucuronidase | Aspergillus niger | Morana[63] |
| Soyasapogenol B | Soyasaponin Ⅰ | Soybean saponin hydrolase | Aspergillus oryzae | Kudou[64] |
| Soyasapogenol B | Soyasaponin | Soybean saponin hydrolase | Neocosmospora vasinfecta | Watanabe[65] |
| Dioscin | Protodioscin | β-Glucosidase | Costus speciosus | Inoue[66] |
| Progenin Ⅲ | Protodioscin | Protodioscin-glycosidase | Aspergillus oryzae | Liu[67] |
| Progenin Ⅴ | Dioscin | α-L-Rhamnosidase | Curvularia lunata | Feng[68] |
| Diosgenyl-glucoside | Dioscin | α-L-Rhamnosidase | Bovine liver | Qian[69] |
| Diosgenin | Dioscin | Dioscin-glycosidase | Absidia | Fu[70] |
| GAMG: Glycyrrhetic acid mono-glucuronide. | ||||
表选项
2.2 黄酮苷的生物转化 黄酮类化合物是植物体内广泛分布的多酚类物质,多以糖苷形式存在。研究发现,具有生物活性的黄酮类化合物是食用植物中最重要的有效成分,具有护肝、抗氧化、抗肿瘤、抗病毒等多种药理学活性[71-72],其活性与结构密切相关[73]。由于大部分黄酮苷难以通过小肠壁进入到血液中,生物利用度低,因此对天然黄酮化合物进行结构修饰成为目前研究的热点。利用糖苷水解酶水解黄酮苷的糖基成为一种提高黄酮化合物活性的有效途径(表 3)。常见的黄酮苷包括芦丁、橙皮苷和柚皮苷,它们的糖基部分通常为芸香糖(α-1, 6连接的鼠李糖和葡萄糖)和新橙皮糖(α-1, 2连接的鼠李糖和葡萄糖),因此糖苷水解酶对其进行水解修饰的方式主要包括外切和内切两种。从Aspergillus niger[74]和Aspergillus nidulans[75]中分离纯化出能水解α-1, 2和α-1, 6鼠李糖苷键的α-鼠李糖苷酶,该酶能水解芦丁、柚皮苷和橙皮苷,分别生成异栎素、洋李苷和橙皮素葡萄糖苷。从Aspergillus aculeatus[76]和Clostridium stercorarium[77]中克隆得到的重组α-鼠李糖苷酶也具有水解黄酮苷中鼠李糖的活性。对于水解上述三种黄酮苷,除了外切糖苷酶外,内切糖苷酶也有大量研究报道。从Penicillium rugulosum[78]、Penicillium decumben[79]和Fagopyri herba[80]中分离纯化出的二糖苷酶,以及从Aspergillus niger[81]中克隆获得的重组芦丁酶,均可水解芦丁生成抗氧化活性更好的槲皮素。从Aspergillus niger BCC 25166[82]中可分离纯化出能水解柚皮苷生成柚皮苷元的柚皮苷酶,Aspergillus niger 1344[83]中的柚皮苷酶可同时水解柚皮苷和芦丁,分别生成柚皮苷元和槲皮素,但不能水解橙皮苷。而Acremonium sp. DSM24697[84]和Actinoplanes missouriensis[85]中的二糖苷酶能水解橙皮苷中的新橙皮糖,生成高活性的橙皮素产物。
表 3. 糖苷水解酶转化黄酮苷 Table 3. Biotransformation of flavonoid glycosides by glycosidase
| Product | Substrate | Reaction | Organism | Reference |
| Isoquercitrin, Prunin, | Rutin, Naringin, | α-Rhamnosidase | Aspergillus niger | Manzanares[74] |
| Hesperetin glucoside | Hesperidin | α-Rhamnosidase | Aspergillus nidulans | Manzanares[75] |
| α-Rhamnosidase | Aspergillus aculeatus | Manzanares[76] | ||
| α-Rhamnosidase | Clostridium stercorarium | Kaur[77] | ||
| Quercetin | Rutin | β-Rutinosidase | Penicillium rugulosum | Narikawa[78] |
| Quercetin | Rutin | β-Glycosidase | Penicillium decumbens | Mamma[79] |
| Quercetin | Rutin | β-Heterodisaccharidase | Fagopyri herba | Baumgertel[80] |
| Quercetin | Rutin | β-Rutinosidase | Aspergillus niger | ?im?lková[81] |
| Naringenin | Naringin | Naringinase | Aspergillus niger | Thammawat[82] |
| Naringenin, Quercetin | Naringin, Rutin | Naringinase | Aspergillus niger | Puri[83] |
| Hesperetin | Hesperidin | Diglycosidase | Acremonium | Pi?uel[84] |
| Hesperetin | Hesperidin | Diglycosidase | Actinoplanes missouriensis | Neher[85] |
| Daidzein | Daidzin | β-Glucosidase | Unculturable microbes | Li[86] |
| Daidzein | Daidzin | β-Glucosidase | Sulfolobus solfataricus | Kim[87] |
| Daidzein | Daidzin | β-Glucosidase | Aspergillus oryzae | Horri[88] |
| Daidzein | Daidzin | β-Glucosidase | Pyrococcus furiosus | Yeom[89] |
| Daidzein, Genistein | Daidzin, Genistin | β-Glucosidase | Bacillus subtilis | Xue[90] |
| β-Glucosidase | Thermotoga maritima | Kuo[91] | ||
| Daidzein, Genistein, | Daidzin, Genistin, | β-Glucosidase | Dalbergia | Chuankhayan[92] |
| Glycitein | Glycitin | β-Glucosidase | Bacteroides thetaiotaomicron | Byun[93] |
| Baicalein | Baicalin | β-Glucuronidase | Scutellaria viscidula | Zhang[94] |
| Tilianin | Linarin | Naringinase | Penicillium decumbens | Cui[95] |
| Butin | Butrin | β-Glucosidase | Almond | Jassbi[96] |
| Phloretin | Phlorizin | β-Glycosidase | Sheep small intestine | Day[97] |
表选项
异黄酮是黄酮类化合物的一种,主要存在于豆科植物中,有助于疾病的预防和人类健康。大豆异黄酮的主要成分为大豆苷、大豆苷元、染料木苷、染料木素、黄豆黄素和黄豆黄素苷元,其中去糖基化的苷元具有更好的生物活性。从红树林土壤的基因文库[86]、Sulfolobus solfataricus[87]、Aspergillus oryzae[88]和Pyrococcus furiosus[89]中克隆获得能水解大豆苷生成大豆苷元的重组β-葡萄糖苷酶;从Thermotoga maritima[90]和Bacillus subtilis[91]中克隆得到的重组β-葡萄糖苷酶能水解大豆苷和染料木苷生成大豆苷元和染料木素;从Dalbergia[92]中分离纯化和从Bacteroides thetaiotaomicron[93]中克隆重组的糖苷酶可以水解大豆苷、染料木苷和黄豆黄素生成大豆苷元、染料木素和黄豆黄素苷元。
糖苷水解酶在其他黄酮苷水解中也有报道和应用(表 3)。研究表明,黄芩苷具有抗肿瘤、抗感染等功效。从Scutellaria viscidula Bge[94]中分离纯化的β-葡萄糖苷酶能够水解黄芩苷生成黄芩素,去糖基化的产物黄芩素具有更好的药理学活性。田蓟苷也是一种稀有黄酮苷,具有抗高血压和镇静等活性,但难以通过直接提取和化学合成的方式获得。Cui等[95]使用柚皮苷酶可将蒙花苷中的鼠李糖水解,生成田蓟苷。此外,Jassbi等[96]使用β-葡萄糖苷酶将紫铆苷水解生成紫铆亭,抗氧化实验结果表明去糖基化的紫铆亭比紫铆苷具有更好的活性。Day等[97]从羊小肠中分离纯化的糖苷酶则可以水解根皮苷生成根皮素。
2.3 其他氧糖苷的生物转化 除了皂苷和黄酮苷外,糖苷水解酶也被应用于水解修饰其他氧糖苷(表 4)。栀子果实是一种传统的中药,用于治疗心脑血管、肝胆等疾病。栀子果实中存在大量京尼平苷,但有效成分是京尼平苷的去糖基化产物京尼平,含量不足0.01%。从Penicillium nigricans[98]和Aspergillus niger[99]中分离纯化出的β-葡萄糖苷酶能转化京尼平苷制备京尼平,以满足大量获取京尼平的需求。牛蒡子具有预防或治疗慢性肾功能衰竭等功效,其有效成分为牛蒡子苷和牛蒡子苷元。Grifola frondosa[100]和Rhizoctonia solani[101]中的β-葡萄糖苷酶能够转化牛蒡子生成牛蒡子苷元。Liu等[102]使用商品化β-葡萄糖苷酶也可以将牛蒡子完全水解获得牛蒡子苷元产物。牛蒡子苷转化为牛蒡子苷元后能有效提高生物利用度。白藜芦醇具有预防肿瘤和动脉粥样硬化等功能,从Aspergillus oryzae sp. 100[103]和Lactobacillus kimchi[104]中分离纯化的β-葡萄糖苷酶,以及Mai等[105]从红树林土壤的宏基因组中克隆获得的重组β-葡萄糖苷酶能够转化虎杖苷生成白藜芦醇。紫杉醇为红豆杉植物的次生代谢产物,对卵巢癌和乳腺癌等具有良好的治疗效果。豆杉中紫杉醇的干重仅为0.02%,而作为废物丢弃的7-木糖-10-去乙酰紫杉醇的含量则是紫杉醇的10倍以上。Dou等[106]利用Cellulosimicrobiumcellulans strain F16分泌的胞外木糖苷酶将7-木糖-10-去乙酰紫杉醇转化成10-去乙酰紫杉醇,再通过一步酰化反应即可生成紫杉醇。
表 4. 糖苷水解酶转化其他氧糖苷 Table 4. Biotransformation of other O-glycosides by glycosidase
| Product | Substrate | Reaction | Organism | Reference |
| Genipin | Geniposide | β-Glucosidase | Penicillium nigricans | Xu[98] |
| Genipin | Geniposide | β-Glucosidase | Aspergillus niger | Gong[99] |
| Arctigenin | Arctiin | β-Glucosidase | Grifola frondosa | Kim[100] |
| Arctigenin | Arctiin | β-Glucosidase | Rhizoctonia solani | Kuo[101] |
| Arctigenin | Arctiin | β-Glucosidase | Commercial | Liu[102] |
| Resveratrol | Polydatin | β-Glucosidase | Aspergillus oryzae | Chen[103] |
| Resveratrol | Polydatin | β-Glucosidase | Lactobacillus kimchi | Ko[104] |
| Resveratrol | Polydatin | β-Glucosidase | Unculturable microbes | Mai[105] |
| 10-Deacetylpaclitaxel | 7-Xylosyl-10-deacetylpaclitaxel | β-Xylosidase | Cellulosimicrobium cellulans | Dou[106] |
表选项
3 碳糖苷及硫糖苷的生物转化 除了氧糖苷外,糖苷水解酶同样被应用于碳糖苷和硫糖苷的水解修饰研究中(表 5)。碳糖苷是由苷元酚羟基所活化的邻位或对位的氢与糖基脱水缩合而形成。碳苷黄酮具有抗炎抑菌、抗肿瘤、降血糖和增强免疫等多种活性[107]。与氧苷黄酮相比,碳苷黄酮具有更高的稳定性,可能被完整吸收,成为潜在的药物分子[108]。由于碳糖苷键难以被水解,因此碳苷黄酮的水解研究报道较少。Sanugul等[109]从人粪便细菌混合物中分离得到一株细菌,该菌在芒果苷诱导下分泌一种糖苷酶,能水解芒果苷中碳糖苷键,生成活性更好的芒果苷元。Nakamura等[110]从人肠道细菌中分离得到strain PUE,该菌种能分离纯化出一种水解葛根素生成苷元的碳糖苷酶。对于碳糖苷酶编码基因的研究中,Braune等[111]发现Eubacterium cellulosolvens中蛋白编码基因dfgA、dfgB、dfgC、dfgD和dfgE共同表达出能够水解异荭草素生成相应苷元的碳糖苷酶。
表 5. 酶法转化碳糖苷和硫糖苷 Table 5. Biotransformation of C-glycosides and S-glycosides by glycosidase
| Product | Substrate | Reaction | Organism | Reference |
| Norathyriol | Mangiferin | C-glucosyl-cleaving enzyme | Bacteroides | Sanugul[109] |
| Daidzein | Puerarin | C-glucosyl-cleaving enzyme | Human intestinal bacterium | Nakamura[110] |
| Luteolin | Homoorientin | C-glucosyl-cleaving enzyme | Eubacterium cellulosolvens | Braune[111] |
| Sulforaphane | Glucoraphanin | Myrosinase | Broccoli seeds | Shen[114] |
表选项
硫代葡萄糖苷是一类重要的硫糖苷化合物,在十字花科植物中广泛存在,如芥菜、青花菜、大蒜等[112]。研究表明,食用十字花科植物可有效预防乳腺癌、肺癌以及结肠癌等多种癌症,其主要活性成分为硫代葡萄糖苷降解后产生的异硫氰酸酯[113]。硫代葡糖苷酶又称黑芥子酶,主要存在于十字花科植物中,但与硫代葡萄糖苷分布在不同位置。只有当细胞破碎时,它们才会混合发生反应,由于内源黑芥子酶含量较少,难以有效水解硫代葡萄糖苷生成活性产物。萝卜硫素是一种具有药理活性的异硫氰酸酯,Shen等[114]用外源黑芥子酶将萝卜硫苷成功转化为萝卜硫素。目前,对碳糖苷和硫糖苷进行水解修饰的研究较少,未来更多的碳糖苷和硫糖苷的开发及结构修饰将为药物开发提供更多的候选分子。
4 展望 糖苷具有抗炎症、抗氧化和抗肿瘤等多种生物活性,拥有开发药品、保健品和化妆品的良好前景。随着现代生物技术的进步,提取分离及分析测试方法不断改善。酶学的发展开拓了生物转化技术的广泛用途,利用酶转化糖苷越来越受到重视。通过糖苷水解酶制备新的糖苷与苷元,将改变糖苷的生物活性,为药物和保健食品提供丰富的资源。目前对酶催化机制的研究相对薄弱,尤其从分子水平解析酶的立体结构、探究酶的结构与其选择性的关系研究较少,还不够深入。分子生物学与结构生物学等现代生物技术将逐步解决这些科学问题,生物转化制备新的糖苷类化合物将越来越广泛地应用在工农业生产中。
References
| [1] | Gleadow RM, Woodrow IE. Mini-Review:Constraints on effectiveness of cyanogenic glycosides in herbivore defense. Journal of Chemical Ecology, 2002, 28(7): 1301-1313. DOI:10.1023/A:1016298100201 |
| [2] | Moghadamtousi SZ, Kamarudin MNA, Chan CK, Goh BH, Kadir HA. Phytochemistry and biology of Loranthus parasiticus Merr, a commonly used herbal medicine. The American Journal of Chinese Medicine, 2014, 42(1): 23-35. DOI:10.1142/S0192415X14500025 |
| [3] | Mochizuki M, Yoo Y, Matsuzawa K, Sato K, Saiki I, Tonooka S, Samukawa K, Azuma I. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside-Rb2, 20(R)-and 20(S)-ginsenoside-Rg3, of red ginseng. Biological and Pharmaceutical Bulletin, 1995, 18(9): 1197-1202. DOI:10.1248/bpb.18.1197 |
| [4] | Chauhan PS, Satti NK, Suri KA, Amina M, Bani S. Stimulatory effects of Cuminum cyminum and flavonoid glycoside on Cyclosporine-A and restraint stress induced immune-suppression in Swiss albino mice. Chemico-Biological Interactions, 2010, 185(1): 66-72. DOI:10.1016/j.cbi.2010.02.016 |
| [5] | Zhang SZ. Glycobiology:A new frontier of life sciences. Chemistry of Life, 1999, 19(3): 103-106. (in Chinese) 张树政. 糖生物学:生命科学中的新前沿. 生命的化学, 1999, 19(3): 103-106. |
| [6] | Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05×1012 structures for a reducing hexasaccharide:the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology, 1994, 4(6): 759-767. DOI:10.1093/glycob/4.6.759 |
| [7] | Zhao Y, Kang LP, Liu YX, Liang YG, Tan DW, Yu ZY, Cong YW, Ma BP. Steroidal saponins from the rhizome of Paris polyphylla and their cytotoxic activities. Planta Medica, 2009, 75(4): 356-363. DOI:10.1055/s-0028-1088380 |
| [8] | Scigelova M, Singh S, Crout DHG. Glycosidases-a great synthetic tool. Journal of Molecular Catalysis B:Enzymatic, 1999, 6(5): 483-494. DOI:10.1016/S1381-1177(99)00012-0 |
| [9] | Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research, 2014, 42(D1): D490-D495. DOI:10.1093/nar/gkt1178 |
| [10] | Yue HA, Xie YM, Zhang SZ, Fang XF. Studies on the Rhizopus Ⅳ. The amyloglucosidases of Rhizopus. Acta Microbiologica Sinica, 1996, 12(2): 187-193. (in Chinese) 乐华爱, 谢玉梅, 张树政, 方心芳. 根霉的研究Ⅳ.根霉淀粉葡萄糖苷酶的研究. 微生物学报, 1996, 12(2): 187-193. |
| [11] | Yu W, Zhu LX, Liu WF, Jin C, Dong ZY. Isolation, identification and phylogenetic analysis of an endoglycoceramidase producing bacterium. Acta Microbiologica Sinica, 2003, 43(2): 151-155. (in Chinese) 于巍, 祝令香, 刘伟丰, 金城, 董志扬. 一株神经节苷脂内切糖苷酶产生菌的分离、鉴定及系统发育分析. 微生物学报, 2003, 43(2): 151-155. |
| [12] | Peng P, Wu J, Cheng AC, Gao QY, Zhang SZ. Cloning and expression of the α-amylase gene from a Bacillus sp. WS06, and characterization of the enzyme. Acta Microbiologica Sinica, 2005, 45(6): 876-880. (in Chinese) 彭平, 吴襟, 程安春, 高启禹, 张树政. 芽孢杆菌α-淀粉酶基因的克隆、表达和酶学性质分析. 微生物学报, 2005, 45(6): 876-880. |
| [13] | Tang SY, Le QT, Shim JH, Yang SJ, Auh JH, Park C, Park KH. Enhancing thermostability of maltogenic amylase from Bacillus thermoalkalophilus ET2 by DNA shuffling. The FEBS Journal, 2006, 273(14): 3335-3345. DOI:10.1111/ejb.2006.273.issue-14 |
| [14] | Muffler K, Leipold D, Scheller MC, Haas C, Steingroewer J, Bley T, Neuhaus HE, Mirata MA, Schrader J, Ulber R. Biotransformation of triterpenes. Process Biochemistry, 2011, 46(1): 1-15. DOI:10.1016/j.procbio.2010.07.015 |
| [15] | Shin KC, Oh DK. Classification of glycosidases that hydrolyze the specific positions and types of sugar moieties in ginsenosides. Critical Reviews in Biotechnology, 2016, 36(6): 1036-1049. DOI:10.3109/07388551.2015.1083942 |
| [16] | Das S, Rosazza JPN. Microbial and enzymatic transformations of flavonoids. Journal of Natural Products, 2006, 69(3): 499-508. DOI:10.1021/np0504659 |
| [17] | Christensen LP. Ginsenosides:chemistry, biosynthesis, analysis, and potential health effects. Advances in Food and Nutrition Research, 2008, 55: 1-99. DOI:10.1016/S1043-4526(08)00401-4 |
| [18] | Lee JH, Ahn JY, Shin TJ, Choi SH, Lee BH, Hwang SH, Kang JY, Kim HJ, Park CW, Nah SY. Effects of minor ginsenosides, ginsenoside metabolites, and ginsenoside epimers on the growth of Caenorhabditis elegans. Journal of Ginseng Research, 2011, 35(3): 375-383. DOI:10.5142/jgr.2011.35.3.375 |
| [19] | Luan HW, Liu X, Qi XH, Hu Y, Hao DC, Cui Y, Yang L. Purification and characterization of a novel stable ginsenoside Rb1-hydrolyzing β-D-glucosidase from China white jade snail. Process Biochemistry, 2006, 41(9): 1974-1980. DOI:10.1016/j.procbio.2006.04.011 |
| [20] | Son JW, Kim HJ, Oh DK. Ginsenoside Rd production from the major ginsenoside Rb1 by β-glucosidase from Thermus caldophilus. Biotechnology Letters, 2008, 30(4): 713-716. DOI:10.1007/s10529-007-9590-4 |
| [21] | Kim SJ, Lee CM, Kim MY, Yeo YS, Yoon SH, Kang HC, Koo BS. Screening and characterization of an enzyme with β-glucosidase activity from environmental DNA. Journal of Microbiology and Biotechnology, 2007, 17(6): 905-912. |
| [22] | Zhao LG, Xie JC, Zhang XS, Cao FL, Pei JJ. Overexpression and characterization of a glucose-tolerant β-glucosidase from Thermotoga thermarum DSM 5069T with high catalytic efficiency of ginsenoside Rb1 to Rd. Journal of Molecular Catalysis B:Enzymatic, 2013, 95: 62-69. DOI:10.1016/j.molcatb.2013.05.027 |
| [23] | Jung IH, Lee JH, Hyun YJ, Kim DH. Metabolism of ginsenoside Rb1 by human intestinal microflora and cloning of its metabolizing β-D-glucosidase from Bifidobacterium longum H-1. Biological and Pharmaceutical Bulletin, 2012, 35(4): 573-581. DOI:10.1248/bpb.35.573 |
| [24] | Hong H, Cui CH, Kim JK, Jin FX, Kim SC, Im WT. Enzymatic biotransformation of ginsenoside Rb1 and gypenoside ⅩⅦ into ginsenosides Rd and F2 by recombinant β-glucosidase from Flavobacterium johnsoniae. Journal of Ginseng Research, 2012, 36(4): 418-424. DOI:10.5142/jgr.2012.36.4.418 |
| [25] | Shin KC, Seo MJ, Oh HJ, Oh DK. Highly selective hydrolysis for the outer glucose at the C-20 position in ginsenosides by β-glucosidase from Thermus thermophilus and its application to the production of ginsenoside F2 from gypenoside ⅩⅦ. Biotechnology Letters, 2014, 36(6): 1287-1293. DOI:10.1007/s10529-014-1472-y |
| [26] | Zhang CZ, Yu HS, Bao YM, An LJ, Jin FX. Purification and characterization of ginsenoside-α-arabinofuranase hydrolyzing ginsenoside Rc into Rd from the fresh root of Panax ginseng. Process Biochemistry, 2002, 37(7): 793-798. DOI:10.1016/S0032-9592(01)00275-8 |
| [27] | Liu QM, Jung HM, Cui CH, Sung BH, Kim JK, Kim SG, Lee ST, Kim SC, Im WT. Bioconversion of ginsenoside Rc into Rd by a novel α-L-arabinofuranosidase, Abf22-3 from Leuconostoc sp. 22-3:cloning, expression, and enzyme characterization. Antonie van Leeuwenhoek, 2013, 103(4): 747-754. DOI:10.1007/s10482-012-9856-2 |
| [28] | Shin HY, Lee JH, Lee JY, Han YO, Han MJ, Kim DH. Purification and characterization of ginsenoside Ra-hydrolyzing β-D-xylosidase from Bifidobacterium breve K-110, a human intestinal anaerobic bacterium. Biological and Pharmaceutical Bulletin, 2003, 26(8): 1170-1173. DOI:10.1248/bpb.26.1170 |
| [29] | Lee JH, Hyun YJ, Kim DH. Cloning and characterization of α-L-arabinofuranosidase and bifunctional α-L-arabinopyranosidase/β-D-galactopyranosidase from Bifidobacterium longum H-1. Journal of Applied Microbiology, 2011, 111(5): 1097-1107. DOI:10.1111/jam.2011.111.issue-5 |
| [30] | Shin KC, Lee GW, Oh DK. Production of ginsenoside Rd from ginsenoside Rc by α-L-arabinofuranosidase from Caldicellulosiruptor saccharolyticus. Journal of Microbiology and Biotechnology, 2013, 23(4): 483-488. DOI:10.4014/jmb |
| [31] | An DS, Cui CH, Sung BH, Yang HC, Kim SC, Lee ST, Im WT, Kim SG. Characterization of a novel ginsenoside-hydrolyzing α-L-arabinofuranosidase, AbfA, from Rhodanobacter ginsenosidimutans Gsoil 3054T. Applied Microbiology and Biotechnology, 2012, 94(3): 673-682. DOI:10.1007/s00253-011-3614-7 |
| [32] | Yu HS, Liu QM, Zhang CZ, Lu MC, Fu YY, Im WT, Lee ST, Jin FX. A new ginsenosidase from Aspergillus strain hydrolyzing 20-O-multi-glycoside of PPD ginsenoside. Process Biochemistry, 2009, 44(7): 772-775. DOI:10.1016/j.procbio.2009.02.005 |
| [33] | Yan Q, Zhou W, Li XW, Feng MQ, Zhou P. Purification method improvement and characterization of a novel ginsenoside-hydrolyzing β-glucosidase from Paecilomyces Bainier sp. 229. Bioscience, Biotechnology, and Biochemistry, 2008, 72(2): 352-359. DOI:10.1271/bbb.70425 |
| [34] | Quan LH, Min JW, Yang DU, Kim YJ, Yang DC. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant β-glucosidase from Microbacterium esteraromaticum. Applied Microbiology and Biotechnology, 2012, 94(2): 377-384. DOI:10.1007/s00253-011-3861-7 |
| [35] | Quan LH, Wang C, Jin Y, Wang TR, Kim YJ, Yang DC. Isolation and characterization of novel ginsenoside-hydrolyzing glycosidase from Microbacterium esteraromaticum that transforms ginsenoside Rb2 to rare ginsenoside 20(S)-Rg3. Antonie van Leeuwenhoek, 2013, 104(1): 129-137. DOI:10.1007/s10482-013-9933-1 |
| [36] | Du J, Cui CH, Park SC, Kim JK, Yu HS, Jin FX, Sun CK, Kim SC, Im WT. Identification and characterization of a ginsenoside-transforming β-glucosidase from Pseudonocardia sp. Gsoil 1536 and its application for enhanced production of minor ginsenoside Rg2(S). PLoS One, 2014, 9(6): e96914. DOI:10.1371/journal.pone.0096914 |
| [37] | Wang L, Liu QM, Sung BH, An DS, Lee HG, Kim SG, Kim SC, Lee ST, Im WT. Bioconversion of ginsenosides Rb1, Rb2, Rc and Rd by novel β-glucosidase hydrolyzing outer 3-O glycoside from Sphingomonas sp. 2F2:cloning, expression, and enzyme characterization. Journal of Biotechnology, 2011, 156(2): 125-133. DOI:10.1016/j.jbiotec.2011.07.024 |
| [38] | Shin KC, Oh DK. Characterization of a novel recombinant β-glucosidase from Sphingopyxis alaskensis that specifically hydrolyzes the outer glucose at the C-3 position in protopanaxadiol-type ginsenosides. Journal of Biotechnology, 2014, 172: 30-37. DOI:10.1016/j.jbiotec.2013.11.026 |
| [39] | An DS, Cui CH, Lee HG, Wang L, Kim SC, Lee ST, Jin FX, Yu HS, Chin YW, Lee HK, Im WT, Kim SG. Identification and characterization of a novel Terrabacter ginsenosidimutans sp. nov. β-Glucosidase that transforms ginsenoside Rb1 into the rare gypenosides ⅩⅦ and LXXV. Applied and Environmental Microbiology, 2010, 76(17): 5827-5836. DOI:10.1128/AEM.00106-10 |
| [40] | Hou JG, Xue JJ, Sun MQ, Wang CY, Liu L, Zhang DL, Lee MR, Gu LJ, Wang CL, Wang YB, Zheng Y, Li W, Sung CK. Highly selective microbial transformation of major ginsenoside Rb1 to gypenoside LXXV by Esteya vermicola CNU120806. Journal of Applied Microbiology, 2012, 113(4): 807-814. DOI:10.1111/jam.2012.113.issue-4 |
| [41] | Park MK, Cui CH, Park SC, Park SK, Kim JK, Jung MS, Jung SC, Kim SC, Im WT. Characterization of recombinant β-glucosidase from Arthrobacter chlorophenolicus and biotransformation of ginsenosides Rb1, Rb2, Rc, and Rd. Journal of Microbiology, 2014, 52(5): 399-406. DOI:10.1007/s12275-014-3601-7 |
| [42] | Park SY, Bae EA, Sung JH, Lee SK, Kim DH. Purification and characterization of ginsenoside Rb1-metabolizing β-glucosidase from Fusobacterium K-60, a human intestinal anaerobic bacterium. Bioscience, Biotechnology, and Biochemistry, 2001, 65(5): 1163-1169. DOI:10.1271/bbb.65.1163 |
| [43] | Fu Y, Yin ZH, Wu LP, Yin CR. Biotransformation of ginsenoside Rb1 to ginsenoside C-K by endophytic fungus Arthrinium sp. GE 17-18 isolated from Panax ginseng. Letters in Applied Microbiology, 2016, 63(3): 196-201. DOI:10.1111/lam.2016.63.issue-3 |
| [44] | Noh KH, Oh DK. Production of the rare ginsenosides compound K, compound Y, and compound Mc by a thermostable β-glycosidase from Sulfolobus acidocaldarius. Biological and Pharmaceutical Bulletin, 2009, 32(11): 1830-1835. DOI:10.1248/bpb.32.1830 |
| [45] | Liu CY, Jin YH, Yu HS, Sun CK, Gao P, Xiao YK, Zhang TY, Xu LQ, Im WT, Jin FX. Biotransformation pathway and kinetics of the hydrolysis of the 3-O-and 20-O-multi-glucosides of PPD-type ginsenosides by ginsenosidase type Ⅰ. Process Biochemistry, 2014, 49(5): 813-820. DOI:10.1016/j.procbio.2014.02.011 |
| [46] | Quan LH, Min JW, Jin Y, Wang C, Kim YJ, Yang DC. Enzymatic biotransformation of ginsenoside Rb1 to compound K by recombinant β-glucosidase from Microbacterium esteraromaticum. Journal of Agricultural and Food Chemistry, 2012, 60(14): 3776-3781. DOI:10.1021/jf300186a |
| [47] | Gao J, Hu YB, Ji L, Wang N, Wang J, Tai GH, Zhou YF. A novel ginsenoside-hydrolyzing enzyme from Penicillium oxalicum and its application in ginsenoside Rd production. Biocatalysis and Biotransformation, 2013, 31(6): 305-312. DOI:10.3109/10242422.2013.857316 |
| [48] | Gao J, Zhao XS, Liu HB, Fan YY, Cheng HR, Liang F, Chen XX, Wang N, Zhou YF, Tai GH. A highly selective ginsenoside Rb1-hydrolyzing β-D-glucosidase from Cladosporium fulvum. Process Biochemistry, 2010, 45(6): 897-903. DOI:10.1016/j.procbio.2010.02.016 |
| [49] | Yuan Y, Hu YB, Hu CX, Leng JY, Chen HL, Zhao XS, Gao J, Zhou YF. Overexpression and characterization of a glycoside hydrolase family 1 enzyme from Cellulosimicrobium cellulans sp. 21 and its application for minor ginsenosides production. Journal of Molecular Catalysis B:Enzymatic, 2015, 120: 60-67. DOI:10.1016/j.molcatb.2015.06.015 |
| [50] | Quan LH, Min JW, Sathiyamoorthy S, Yang DU, Kim YJ, Yang DC. Biotransformation of ginsenosides Re and Rg1 into ginsenosides Rg2 and Rh1 by recombinant β-glucosidase. Biotechnology Letters, 2012, 34(5): 913-917. DOI:10.1007/s10529-012-0849-z |
| [51] | Cui CH, Liu QM, Kim JK, Sung BH, Kim SG, Kim SC, Im WT. Identification and characterization of a Mucilaginibacter sp. strain QM49β-glucosidase and its use in the production of the pharmaceutically active minor ginsenosides (S)-Rh1 and (S)-Rg2. Applied and Environmental Microbiology, 2013, 79(19): 5788-5798. DOI:10.1128/AEM.01150-13 |
| [52] | Oh HJ, Shin KC, Oh DK. Production of ginsenosides Rg1 and Rh1 by hydrolyzing the outer glycoside at the C-6 position in protopanaxatriol-type ginsenosides using β-glucosidase from Pyrococcus furiosus. Biotechnology Letters, 2014, 36(1): 113-119. DOI:10.1007/s10529-013-1331-2 |
| [53] | Ruan CC, Zhang H, Zhang LX, Liu Zhi, Sun GZ, Lei J, Qin YX, Zheng YN, Li X, Pan HY. Biotransformation of ginsenoside Rf to Rh1 by recombinant β-glucosidase. Molecules, 2009, 14(6): 2043-2048. DOI:10.3390/molecules14062043 |
| [54] | Yu HS, Gong JM, Zhang CZ, Jin FX. Purification and characterization of ginsenoside-α-L-rhamnosidase. Chemical and Pharmaceutical Bulletin, 2002, 50(2): 175-178. DOI:10.1248/cpb.50.175 |
| [55] | Shin KC, Seo MJ, Oh DK. Characterization of β-xylosidase from Thermoanaerobacterium thermosaccharolyticum and its application to the production of ginsenosides Rg1 and Rh1 from notoginsenosides R1 and R2. Biotechnology Letters, 2014, 36(11): 2275-2281. DOI:10.1007/s10529-014-1604-4 |
| [56] | Kim YS, Yoo MH, Lee GW, Choi JG, Kim KR, Oh DK. Ginsenoside F1 production from ginsenoside Rg1 by a purified β-glucosidase from Fusarium moniliforme var. subglutinans. Biotechnology Letters, 2011, 33(12): 2457-2461. DOI:10.1007/s10529-011-0719-0 |
| [57] | Wei Y, Zhao WQ, Zhang Q, Zhao YQ, Zhang YX. Purification and characterization of a novel and unique ginsenoside Rg1-hydrolyzing β-D-glucosidase from Penicillium sclerotiorum. Acta Biochimica et Biophysica Sinica, 2011, 43(3): 226-231. DOI:10.1093/abbs/gmr001 |
| [58] | Kim JK, Cui CH, Yoon MH, Kim SC, Im WT. Bioconversion of major ginsenosides Rg1 to minor ginsenoside F1 using novel recombinant ginsenoside hydrolyzing glycosidase cloned from Sanguibacter keddieii and enzyme characterization. Journal of Biotechnology, 2012, 161(3): 294-301. DOI:10.1016/j.jbiotec.2012.06.021 |
| [59] | Zhao XS, Gao J, Song CC, Fang Q, Wang N, Zhao TJ, Liu DB, Zhou YF. Fungal sensitivity to and enzymatic deglycosylation of ginsenosides. Phytochemistry, 2012, 78: 65-71. DOI:10.1016/j.phytochem.2012.02.027 |
| [60] | Gao J, Hu YB, Meng Y, Meng FL, Guo XQ, Wang N, Wei M, Zhou YF. Simple and efficient preparation of ginsenoside (S)-Rg2 from ginsenoside Re by biotransformation with Cellulosimicrobium sp. 21. Biocatalysis and Biotransformation, 2015, 33(1): 51-60. DOI:10.3109/10242422.2015.1018192 |
| [61] | Park HY, Kim NY, Han MJ, Bae EA, Kim DH. Purification and characterization of two novel β-D-glucuronidases converting glycyrrhizin to 18β-glycyrrhetinic acid-3-O-β-Dglucuronide from Streptococcus LJ-22. Journal of Microbiology and Biotechnology, 2005, 15(4): 792-799. |
| [62] | Zou SP, Liu GY, Kaleem I, Li C. Purification and characterization of a highly selective glycyrrhizin-hydrolyzing β-glucuronidase from Penicillium purpurogenum Li-3. Process Biochemistry, 2013, 48(2): 358-363. DOI:10.1016/j.procbio.2012.12.008 |
| [63] | Morana A, Di Lazzaro A, Di Lernia I, Ponzone C, De Rose M. Enzymatic production of 18-β-glycyrrhetinic acid from Glycyrrhizaglabra L. Biotechnology Letters, 2002, 24(22): 1907-1911. DOI:10.1023/A:1020904325906 |
| [64] | Kudou S, Tsuizaki I, Uchida T, Okubo K. Purification and some properties of soybean saponin hydrolase from Aspergillus oryzae KO-2. Agricultural and Biological Chemistry, 1991, 55(1): 31-36. |
| [65] | Watanabe M, Sumida N, Yanai K, Murakami T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Applied and Environmental Microbiology, 2004, 70(2): 865-872. DOI:10.1128/AEM.70.2.865-872.2004 |
| [66] | Inoue K, Ebizuka Y. Purification and characterization of furostanol glycoside 26-O-β-glucosidase from Costus speciosus rhizomes. FEBS Letters, 1996, 378(2): 157-160. DOI:10.1016/0014-5793(95)01447-0 |
| [67] | Liu TQ, Yu HS, Liu CY, Wang YH, Tang MQ, Yuan XD, Luo N, Wang QY, Xu XD, Jin FX. Protodioscin-glycosidase-1 hydrolyzing 26-O-β-D-glucoside and 3-O-(1→4)-α-Lrhamnoside of steroidal saponins from Aspergillus oryzae. Applied Microbiology and Biotechnology, 2013, 97(23): 10035-10043. DOI:10.1007/s00253-013-4791-3 |
| [68] | Feng B, Hu W, Ma BP, Wang YZ, Huang HZ, Wang SQ, Qian XH. Purification, characterization, and substrate specificity of a glucoamylase with steroidal saponin-rhamnosidase activity from Curvularia lunata. Applied Microbiology and Biotechnology, 2007, 76(6): 1329-1338. DOI:10.1007/s00253-007-1117-3 |
| [69] | Qian S, Wang HY, Zhang CZ, Yu HS. Isolation and characterization of dioscin-α-L-rhamnosidase from bovine liver. Journal of Molecular Catalysis B:Enzymatic, 2013, 97: 31-35. DOI:10.1016/j.molcatb.2013.07.007 |
| [70] | Fu YY, Yu HS, Tang SH, Hu XC, Wang YH, Liu B, Yu CX, Jin FX. New dioscin-glycosidase hydrolyzing multi-glycosides of dioscin from Absidia strain. Journal of Microbiology and Biotechnology, 2010, 20(6): 1011-1017. DOI:10.4014/jmb |
| [71] | Heim KE, Tagliaferro AR, Bobilya DJ. Flavonoid antioxidants:chemistry, metabolism and structure-activity relationships. The Journal of Nutritional Biochemistry, 2002, 13(10): 572-584. DOI:10.1016/S0955-2863(02)00208-5 |
| [72] | Meda AL, Lamien CE, Romito M, Millogo JF, Nacoulma OG. Determination of the total phenolic, flavonoid and proline contents in Burkina Fasan honey, as well as their radical scavenging activity. Food Chemistry, 2005, 91(3): 571-577. DOI:10.1016/j.foodchem.2004.10.006 |
| [73] | Matsuda H, Morikawa T, Toguchida I, Yoshikawa M. Structural requirements of flavonoids and related compounds for aldose reductase inhibitory activity. Chemical and Pharmaceutical Bulletin, 2002, 50(6): 788-795. DOI:10.1248/cpb.50.788 |
| [74] | Manzanares P, de Graaff LH, Visser J. Purification and characterization of an α-L-rhamnosidase from Aspergillus niger. FEMS Microbiology Letters, 1997, 157(2): 279-283. DOI:10.1016/S0378-1097(97)00487-4 |
| [75] | Manzanares P, Orejas M, Iba?ez E, Vallés S, Ramón D. Purification and characterization of an α-L-rhamnosidase from Aspergillus nidulans. Letters in Applied Microbiology, 2000, 31(3): 198-202. DOI:10.1046/j.1365-2672.2000.00788.x |
| [76] | Manzanares P, van den Broeck HC, de Graaff LH, Visser J. Purification and characterization of two different α-L-rhamnosidases, RhaA and RhaB, from Aspergillus aculeatus. Applied and Environmental Microbiology, 2001, 67(5): 2230-2234. DOI:10.1128/AEM.67.5.2230-2234.2001 |
| [77] | Kaur A, Singh S, Singh RS, Schwarz WH, Puri M. Hydrolysis of citrus peel naringin by recombinant α-L-rhamnosidase from Clostridium stercorarium. Journal of Chemical Technology and Biotechnology, 2010, 85(10): 1419-1422. DOI:10.1002/jctb.v85:10 |
| [78] | Narikawa T, Shinoyama H, Fujii T. A β-rutinosidase from Penicillium rugulosum IFO 7242 that is a peculiar flavonoid glycosidase. Bioscience, Biotechnology, and Biochemistry, 2000, 64(6): 1317-1319. DOI:10.1271/bbb.64.1317 |
| [79] | Mamma D, Kalogeris E, Hatzinikolaou DG, Lekanidou A, Kekos D, Macris BJ, Christakopoulos P. Biochemical characterization of the multi-enzyme system produced by Penicillium decumbens grown on rutin. Food Biotechnology, 2004, 18(1): 1-18. DOI:10.1081/FBT-120030382 |
| [80] | Baumgertel A, Grimm R, Eisenbei? W, Kreis W. Purification and characterization of a flavonol 3-O-β-heterodisaccharidase from the dried herb of Fagopyrum esculentum Moench. Phytochemistry, 2003, 64(2): 411-418. DOI:10.1016/S0031-9422(03)00418-7 |
| [81] | ?im?íková D, Kotik M, Weignerová L, Halada P, Pelantová H, Adamcová K, K?en V. α-L-rhamnosyl-β-D-glucosidase (rutinosidase) from Aspergillus niger:characterization and synthetic potential of a novel diglycosidase. Advanced Synthesis & Catalysis, 2015, 357(1): 107-117. |
| [82] | Thammawat K, Pongtanya P, Juntharasri V, Wongvithoonyaporn P. Isolation, preliminary enzyme characterization and optimization of culture parameters for production of naringinase isolated from Aspergillus niger BCC 25166. Kasetsart Journal (Natural Science), 2008, 42(1): 61-72. |
| [83] | Puri M, Kalra S. Purification and characterization of naringinase from a newly isolated strain of Aspergillus niger 1344 for the transformation of flavonoids. World Journal of Microbiology and Biotechnology, 2005, 21(5): 753-758. DOI:10.1007/s11274-004-5488-7 |
| [84] | Pi?uel L, Breccia JD, Guisán JM, López-Gallego F. Production of hesperetin using a covalently multipoint immobilized diglycosidase from Acremonium sp. DSM24697. Journal of Molecular Microbiology and Biotechnology, 2013, 23(6): 410-417. DOI:10.1159/000353208 |
| [85] | Neher BD, Mazzaferro LS, Kotik M, Oyhenart J, Halada P, K?en V, Breccia JD. Bacteria as source of diglycosidase activity:Actinoplanes missouriensis produces 6-O-α-L-rhamnosyl-β-d-glucosidase active on flavonoids. Applied Microbiology and Biotechnology, 2016, 100(7): 3061-3070. DOI:10.1007/s00253-015-7088-x |
| [86] | Li G, Jiang Y, Fan XJ, Liu YH. Molecular cloning and characterization of a novel β-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library. Bioresource Technology, 2012, 123: 15-22. DOI:10.1016/j.biortech.2012.07.083 |
| [87] | Kim BN, Yeom SJ, Kim YS, Oh DK. Characterization of a β-glucosidase from Sulfolobus solfataricus for isoflavone glycosides. Biotechnology Letters, 2012, 34(1): 125-129. DOI:10.1007/s10529-011-0739-9 |
| [88] | Horii K, Adachi T, Matsuda T, Tanaka T, Sahara H, Shibasaki S, Ogino C, Hata Y, Ueda M, Kondo A. Improvement of isoflavone aglycones production using β-glucosidase secretory produced in recombinant Aspergillus oryzae. Journal of Molecular Catalysis B:Enzymatic, 2009, 59(4): 297-301. DOI:10.1016/j.molcatb.2008.11.013 |
| [89] | Yeom SJ, Kim BN, Kim YS, Oh DK. Hydrolysis of isoflavone glycosides by a thermostable β-glucosidase from Pyrococcus furiosus. Journal of Agricultural and Food Chemistry, 2012, 60(6): 1535-1541. DOI:10.1021/jf204432g |
| [90] | Xue YM, Yu JJ, Song XF. Hydrolysis of soy isoflavone glycosides by recombinant β-glucosidase from hyperthermophile Thermotoga maritima. Journal of Industrial Microbiology & Biotechnology, 2009, 36(11): 1401-1408. |
| [91] | Kuo LC, Lee KT. Cloning, expression, and characterization of two β-glucosidases from isoflavone glycoside-hydrolyzing Bacillus subtilis natto. Journal of Agricultural and Food Chemistry, 2008, 56(1): 119-125. DOI:10.1021/jf072287q |
| [92] | Chuankhayan P, Rimlumduan T, Svasti J, Cairns JRK. Hydrolysis of soybean isoflavonoid glycosides by Dalbergia β-glucosidases. Journal of Agricultural and Food Chemistry, 2007, 55(6): 2407-2412. DOI:10.1021/jf062885p |
| [93] | Byun DH, Choi HJ, Lee HW, Jeon HY, Choung WJ, Shim JH. Properties and applications of β-glycosidase from Bacteroides thetaiotaomicron that specifically hydrolyses isoflavone glycosides. International Journal of Food Science and Technology, 2015, 50(6): 1405-1412. DOI:10.1111/ijfs.2015.50.issue-6 |
| [94] | Zhang CZ, Zhang YF, Chen JP, Liang XM. Purification and characterization of baicalin-β-D-glucuronidase hydrolyzing baicalin to baicalein from fresh roots of Scutellaria viscidula Bge. Process Biochemistry, 2005, 40(5): 1911-1915. DOI:10.1016/j.procbio.2004.07.003 |
| [95] | Cui P, Dou TY, Li SY, Lu JX, Zou LW, Wang P, Sun YP, Hao DC, Ge GB. Highly selective and efficient biotransformation of linarin to produce tilianin by naringinase. Biotechnology Letters, 2016, 38(8): 1367-1373. DOI:10.1007/s10529-016-2116-1 |
| [96] | Jassbi AR, Singh P, Krishna V, Gupta PK, Tahara S. Antioxidant study and assignments of NMR spectral data for 3', 4', 7-Trihydroxyflavanone 3', 7-Di-O-β-D-glucopyranoside (Butrin) and its hydrolyzed product. Chemistry of Natural Compounds, 2004, 40(3): 250-253. DOI:10.1023/B:CONC.0000039134.46227.1f |
| [97] | Day AJ, Ca?ada FJ, Díaz JC, Kroon PA, Mclauchlan R, Faulds CB, Plumb GW, Morgan MRA, Williamson G. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Letters, 2000, 468(2/3): 166-170. |
| [98] | Xu MM, Sun Q, Su J, Wang JF, Xu C, Zhang T, Sun QL. Microbial transformation of geniposide in Gardenia jasminoides Ellis into genipin by Penicillium nigricans. Enzyme and Microbial Technology, 2008, 42(5): 440-444. DOI:10.1016/j.enzmictec.2008.01.003 |
| [99] | Gong GH, Zheng ZM, Liu H, Wang L, Diao JS, Wang P, Zhao GH. Purification and characterization of a β-glucosidase from Aspergillus nigerand its application in the hydrolysis of geniposide to genipin. Journal of Microbiology and Biotechnology, 2014, 24(6): 788-794. DOI:10.4014/jmb.1401.01053 |
| [100] | Kim JH, Bae JT, Song MH, Lee GS, Choe SY, Pyo HB. Biological activities of Fructus arctii fermented with the basidiomycete Grifola frondosa. Archives of Pharmacal Research, 2010, 33(12): 1943-1951. DOI:10.1007/s12272-010-1209-y |
| [101] | Kuo PC, Chen ZY, Chen MF. Biopreparation of an anti-inflammatory agent, diarctigenin, from arctiin isolated from Arctium lappa by Rhizoctonia solani AG-4. Tetrahedron Letters, 2013, 54(50): 6955-6958. DOI:10.1016/j.tetlet.2013.10.057 |
| [102] | Liu F, Xi XJ, Wang M, Fan L, Geng YL, Wang X. Isolation and purification of arctigenin from Fructus arctii by enzymatic hydrolysis combined with high-speed counter-current chromatography. Journal of Separation Science, 2014, 37(4): 376-381. DOI:10.1002/jssc.v37.4 |
| [103] | Chen M, Li D, Gao ZQ, Zhang CZ. Enzymatic transformation of polydatin to resveratrol by piceid-β-D-glucosidase from Aspergillus oryzae. Bioprocess and Biosystems Engineering, 2014, 37(7): 1411-1416. DOI:10.1007/s00449-013-1113-1 |
| [104] | Ko JA, Park JY, Kwon HJ, Ryu YB, Jeong HJ, Park SJ, Kim CY, Oh HM, Park CS, Lim YH, Kim D, Rho MC, Lee WS, Kim YM. Purification and functional characterization of the first stilbene glucoside-specific β-glucosidase isolated from Lactobacillus kimchi. Enzyme and Microbial Technology, 2014, 67: 59-66. DOI:10.1016/j.enzmictec.2014.09.001 |
| [105] | Mai ZM, Su HF, Zhang S. Characterization of a metagenome-derived β-glucosidase and its application in conversion of polydatin to resveratrol. Catalysts, 2016, 6(3): 35. DOI:10.3390/catal6030035 |
| [106] | Dou TY, Luan HW, Liu XB, Li SY, Du XF, Yang L. Enzymatic hydrolysis of 7-xylosyltaxanes by an extracellular xylosidase from Cellulosimicrobium cellulans. Biotechnology Letters, 2015, 37(9): 1905-1910. DOI:10.1007/s10529-015-1867-4 |
| [107] | Courts FL, Williamson G. The occurrence, fate and biological activities of C-glycosyl flavonoids in the human diet. Critical Reviews in Food Science and Nutrition, 2015, 55(10): 1352-1367. DOI:10.1080/10408398.2012.694497 |
| [108] | Ghosh RK, Ghosh SM, Chawla S, Jasdanwala SA. SGLT2 inhibitors:a new emerging therapeutic class in the treatment of type 2 diabetes mellitus. The Journal of Clinical Pharmacology, 2012, 52(4): 457-463. DOI:10.1177/0091270011400604 |
| [109] | Sanugul K, Akao T, Li Y, Kakiuchi N, Nakamura N, Hattori M. Isolation of a human intestinal bacterium that transforms mangiferin to norathyriol and inducibility of the enzyme that cleaves a C-glucosyl bond. Biological and Pharmaceutical Bulletin, 2005, 28(9): 1672-1678. DOI:10.1248/bpb.28.1672 |
| [110] | Nakamura K, Komatsu K, Hattori M, Iwashima M. Enzymatic cleavage of the C-glucosidic bond of puerarin by three proteins, Mn2+, and oxidized form of nicotinamide adenine dinucleotide. Biological and Pharmaceutical Bulletin, 2013, 36(4): 635-640. DOI:10.1248/bpb.b12-01011 |
| [111] | Braune AW, Engst W, Blaut M. Identification and functional expression of genes encoding flavonoid O-and C-glycosidases in intestinal bacteria. Environmental Microbiology, 2016, 18(7): 2117-2129. DOI:10.1111/1462-2920.12864 |
| [112] | Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry, 2001, 56(1): 5-51. DOI:10.1016/S0031-9422(00)00316-2 |
| [113] | Tse G, Eslick GD. Cruciferous vegetables and risk of colorectal neoplasms:a systematic review and meta-analysis. Nutrition and Cancer, 2014, 66(1): 128-139. DOI:10.1080/01635581.2014.852686 |
| [114] | Shen LQ, Su GY, Wang XY, Du QZ, Wang KW. Endogenous and exogenous enzymolysis of vegetable-sourced glucosinolates and influencing factors. Food Chemistry, 2010, 119(3): 987-994. DOI:10.1016/j.foodchem.2009.08.003 |
