刘艳如

 , 赵丙春, 董盼盼, 邱黎清, 黄建忠, 朱晓兰, 王作镇, 舒正玉
, 赵丙春, 董盼盼, 邱黎清, 黄建忠, 朱晓兰, 王作镇, 舒正玉

福建师范大学生命科学学院, 工业微生物发酵技术国家地方联合工程研究中心, 教育部工业微生物工程中心, 福建 福州 350108
收稿日期:2016-10-01;修回日期:2016-11-11;网络出版日期:2016-12-02
基金项目:国家自然科学基金(31370802,30870545);福建省科技厅重点项目(2013H0021);福建省自然科学基金****项目(2009J06013)
*通信作者:刘艳如, Tel/Fax:+86-591-22868212, E-mail:yrliu@fjnu.edu.cn
舒正玉, Tel/Fax:+86-591-22868212, E-mail:shuzhengyu@fjnu.edu.cn
摘要: [目的]借助蛋白质工程技术提高伯克霍尔德菌ZYB002脂肪酶LipA的热稳定性,以期更好地将其应用于工业生产中。[方法]利用YASARA、FoldX、Rosetta、Gromacs等生物信息学软件,构建1个脂肪酶LipA的热稳定性提高的微型突变体电子文库;通过对突变体的结构信息和自由能变化进行评估,筛选出潜在的有价值的突变体。继而利用基因定点突变技术,构建上述候选突变体的突变基因文库,通过实验筛选出热稳定性提高的脂肪酶LipA突变体。[结果]利用上述方法,从构建的20个脂肪酶LipA突变体中,筛选到热稳定性有显著提高的3个突变体LipA-Asn125Asp、LipA-Asn125Glu和LipA-Gln262Glu。经55℃处理12 min后,上述3个突变体的T5012值较野生型分别提高4.0℃、5.5℃和4.4℃;在55℃下的半衰期较野生型分别提高了2.2倍、3.8倍和2.6倍。[结论]利用生物信息学软件,构建脂肪酶LipA突变体电子文库,结合蛋白质的结构信息,可以快速筛选到热稳定性提高的脂肪酶LipA突变体。
关键词: 伯克霍尔德菌 脂肪酶A 电子设计与筛选 盐桥 热稳定性突变体
Computer-aid screening of thermostable lipase LipA from Burkholderia sp. ZYB002
Yanru Liu

 , Bingchun Zhao, Panpan Dong, Liqing Qiu, Jianzhong Huang, Xiaolan Zhu, Zuozhen Wang, Zhengyu Shu
, Bingchun Zhao, Panpan Dong, Liqing Qiu, Jianzhong Huang, Xiaolan Zhu, Zuozhen Wang, Zhengyu Shu

National & Local United Engineering Research Center of Industrial Microbiology and Fermentation Technology, Engineering Research Center of Industrial Microbiology, Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou 350108, Fujian Province, China
Received 1 October 2016; Revised 11 November 2016; Published online 2 December 2016
*Corresponding author: Yanru Liu, Tel/Fax:+86-591-22868212, E-mail:yrliu@fjnu.edu.cn
Zhengyu Shu, Tel/Fax:+86-591-22868212, E-mail:shuzhengyu@fjnu.edu.cn
Supported by the National Natural Science Foundation of China (31370802, 30870545), by the Key Project from Science and Technology Bureau of Fujian Province (2013H0021) and by the Natural Science Foundation for Distinguished Young Scholar of Fujian Province (2009J06013)
Abstract: [Objective]We improved the thermostability of lipase LipA from Burkholderia cecapia ZYB002 using protein engineering technology.[Methods]Lipase LipA mutant library was designed and screened using the following software, YASARA, FoldX, Rosetta, and Gromacs. We screened 27 thermostable lipase LipA mutants displaying the salt bridge effect among the resulting library of 341 variants, and then further screened using the site-directed mutagenesis technology.[Results]Three mutants LipA-Asn125Asp, LipA-Asn125Glu and LipA-Gln262Glu displayed improved thermostability. The T5012 value of LipA-Asn125Asp, LipA-Asn125Glu and LipA-Gln262Glu increased by 4.0℃, 5.5℃ and 4.4℃, respectively. The half-life of LipA-Asn125Asp, LipA-Asn125Glu and LipA-Gln262Glu at 55℃ increased by 2.23-fold, 3.8-fold and 2.6-fold, respectively.[Conclusion]It is feasible to screen thermostable mutant from the computationally designed library.
Key words: Burkholderia sp.ZYB002 lipase LipA in silico design and screening salt bridge thermostable mutant
微生物脂肪酶(Triacylglycerol acylhydrolase,EC 3.1.1.3) 又名三酰甘油酯水解酶,是一类能催化长链脂肪酸甘油酯水解为甘油和脂肪酸的生物催化剂。作为一种非水相酶,脂肪酶已被广泛应用到食品加工、精细化工、生物能源及制药等诸多领域,具有重要的经济价值[1]。大量微生物脂肪酶资源陆续被挖掘、开发,以Candida antarctica脂肪酶B、Burkholderia cepacia脂肪酶A和Rhizopus oryzae脂肪酶为代表的系列脂肪酶均已实现了商业化生产[2]。到目前为止,已开发的脂肪酶多为中温脂肪酶,欠佳的热稳定性一定程度上制约了脂肪酶的规模化工业应用。虽然近些年来陆续有嗜脂肪酶或耐热脂肪酶资源的零星报道[3-4],但极端脂肪酶到目前为止,尚未实现工业化生产及大规模工业化应用。
快速获得具有良好热稳定性的突变体酶分子,一直是生物催化剂领域诸多专家(包括学界和业界)的追求目标。近些年来,基于蛋白质工程技术实现快速筛选或构建热稳定的酶分子,得到迅猛发展,诞生了系列新方法新技术。以Reetz教授为代表的科学家基于酶分子氨基酸残基B-factor值的大小作为筛选影响酶分子热稳定性的潜在氨基酸残基突变位点的标准,继而利用迭代饱和突变技术(Iterative saturation mutagenesis),极大地缩小了突变文库容量,可快速筛选出理想的热稳定性突变体[5-6]。随着越来越多蛋白质晶体结构的解析及生物信息学的迅猛发展,通过构建电子文库,并以ΔΔGFold数值高低及突变体结构模型作为筛选标准,能进一步缩小突变文库的数量。Floor等利用此方法,从仅含150个突变体的突变库中快速筛选到表观熔点温度(Apparent melting temperature)提高了23 ℃,60 ℃下半衰期延长了200倍的卤代烷烃脱卤酶(Haloalkane dehalogenase)热稳定性突变体[7]。在酶分子3D结构信息指导下,还可直接理性设计出热稳定性突变体。如Zhou等利用来自于嗜热水解酶的结构模体(motiy)置换中温脂肪酶的相应模体获得的脂肪酶突变体,50 ℃下的温度稳定性提高了100倍[8]。
本课题组自主分离筛选到1株脂肪酶高产菌株,经鉴定并命名为伯克霍尔德菌ZYB002 (Burkholderia sp.)[9]。在前期研究中,该菌株分泌的胞外脂肪酶LipA,在TMP纸浆造纸工艺中,可有效分解纸浆中的树脂成分,对控制树脂障碍具有一定的潜在价值[10]。为进一步提高该脂肪酶的应用效果,有必要利用蛋白质工程技术提高其热稳定性(机械浆温度高达80 ℃)。本文报道利用生物信息学软件设计、筛选潜在的Burkholderia sp. ZYB002脂肪酶热稳定性突变体;通过基因定点突变技术,获得重组突变体蛋白,筛选到性能优良的热稳定性突变体,实验验证了该技术的可行性和有效性。
1 材料和方法 1.1 材料
1.1.1 菌株与载体: Burkholderia sp. ZYB002菌株由本实验室从油污土壤中分离鉴定并保存[9];脂肪酶编码基因lipA及其对应的伴侣蛋白编码基因lipB由本实验室克隆并提交NCBI核酸数据库(登录号为:EU768869)。表达载体pACYC-lipA/lipB及重组表达菌株E. coli BL21(DE3)-pACYC-lipA/lipB由本实验室构建并保存[11]。
1.1.2 工具酶、引物及试剂: PrimeSTAR HS DNA Polymerase、QuickCut Dpn I限制性内切酶及标准分子量的DNA Marker均购于宝生物工程(大连)有限公司;Wizard SV Gel and PCR Clean-Up System试剂盒购于普洛麦格(北京)生物技术有限公司;4-硝基苯月桂酸酯(p-Nitrophenyl laurate,pNPL)购于Sigma (美国)公司;氯霉素购于鼎国生物技术有限公司(北京);其他试剂均为市售分析纯。本实验使用的引物对如表 1。 表 1. 本实验使用的PCR引物对 Table 1. Pairs of the primers used in this study
| Primers | Mutation sites | Oligonucleotide sequence (5'→3’)* | Tm /℃ | Annealing temperature/℃ |
| BF1F | Q34K | CGGCATCAAAGAGGACCTGCAACAG | 65.3 | 60 |
| BF1R | GTCCTCTTTGATGCCGTACCAATACTCGAG | 66.1 | ||
| BF2F | Q39K | GGACCTGCAAAAAAACGGTGCGACC | 65.3 | 60 |
| BF2R | CGCACCGTTTTTTTGCAGGTCCTCC | 65.3 | ||
| BF3F | N40R | CTGCAACAGCGTGGTGCGACCGTCT | 68.5 | 63 |
| BF3R | TCGCACCACGCTGTTGCAGGTCCTC | 68.5 | ||
| BF4F | T79E | GCGACGGGGGCGGAAAAGGTGAATC | 68.5 | 62 |
| BF4F | GCCGACGAGATTCACCTTTTCCGCC | 66.9 | ||
| BF5F | A97K | GCTATGTTAAAGCCGTCGCGCCCGATCTCG | 70.2 | 60 |
| BF5R | GACGGCTTTAACATAGCGCGACGAGAGGCC | 70.2 | ||
| BF6F | A100K | CCGTCAAACCCGATCTCGTTGCGTC | 66.9 | 62 |
| BF6R | CGGGTTTGACGGCAGCAACATAGCG | 66.9 | ||
| BF7F | D102E | GTCGCGCCCGAACTCGTTGCGTCGG | 71.8 | 62 |
| BF7R | CAACGAGTTCGGGCGCGACGGCAGC | 71.8 | ||
| BF8F | N125E | GCCGACTTCGTGCAGGAAGTGCTGG | 68.5 | 63 |
| BF8R | CGCCAGCACTTCCTGCACGAAGTCG | 68.5 | ||
| BF9F | N125D | AGGATGTGCTGGCGTACGATCCGAC | 66.9 | 62 |
| BF9R | ATCGTACGCCAGCACATCCTGCACG | 66.9 | ||
| BF10F | S135K | CGGGCTTCGTTCATCGGTGATCGCC | 68.5 | 63 |
| BF10R | CCGATGAACGAAGCCCGGTCGGATC | 68.5 | ||
| BF11F | A140R | GGTGATCCGTGCGTTCGTCAATGTG | 65.3 | 60 |
| BF11R | ACGAACGCACGGATCACCGATGAAG | 65.3 | ||
| BF12F | N144K | GATCGCCGCGTTCGTCAAAGTGTTC | 65.3 | 62 |
| BF12R | GATTCCGAACACTTTGACGAACGCG | 63.6 | ||
| BF13F | S152K | ACGAGCAAAAGCCACAACACCAACC | 63.6 | 58 |
| BF13R | TTGTGGCTTTTGCTCGTCAGGATTC | 62.0 | ||
| BF14F | T166K | CTGCAGAAACTGACCACCGCACGGG | 68.5 | 63 |
| BF14R | TGGTCAGTTTCTGCAGTGCGGCGAG | 66.9 | ||
| BF15F | Y175D | CCGCCACGGATAACCAGAACTATCC | 65.3 | 60 |
| BF15R | ATAGTTCTGGTTATCCGTGGCGGCC | 65.3 | ||
| BF16F | N178R | TACAACCAGCGTTATCCGAGCGCGG | 66.9 | 62 |
| BF16R | CGGATAACGCTGGTTGTACGTGGCG | 66.9 | ||
| BF17F | V199K | GACCGAAACCAAAGGCGGCAACACG | 66.9 | 62 |
| BF17R | GTGCGTGTTGCCGCCTTTGGTTTCG | 66.9 | ||
| BF18F | Q262E | CTCCGGAGAAAACGACGGGCTCGTG | 68.5 | 63 |
| BF18R | GTCGTTTTCTCCGGAGCCGCGGTTG | 68.5 | ||
| BF19F | L278E | GTACGGCAAGGTGGAAAGCACGAGC | 66.9 | 62 |
| BF19R | TTGTAGCTCGTGCTTTCCACCTTGC | 63.6 | ||
| BF20F | S281D | CAAGGTGCTGAGCACGGATTACAAG | 63.6 | 58 |
| BF20R | GGTTCCACTTGTAATCCGTGCTCAG | 63.6 | ||
| BF21F | N285R | CTACAAGTGGCGTCACCTCGACGAG | 66.9 | 62 |
| BF21R | GAGGTGACGCCACTTGTAGCTCGTG | 66.9 | ||
| BF22F | V305R | GTATGCTGAAGATCCGCGTGCGGTG | 66.9 | 60 |
| BF22R | ATCACCGCACGCGGATCTTCAGCAT | 65.3 | ||
| BF23F | A306E | GATCCGGTCGAAGTGATCCGCACGC | 68.5 | 62 |
| BF23R | GCATGCGTGCGGATCACTTCGACCG | 68.5 | ||
| *Bold nucleotides: the mutated sites. | ||||
表选项
1.2 影响脂肪酶LipA热稳定性的潜在突变位点的筛选 影响脂肪酶LipA热稳定性的潜在突变位点筛选方法如下:(1) 脂肪酶LipA三级结构模型的模拟、优化及评估,具体方法参照刘艳如等[12]。(2) 利用YASARA软件[13],确定距离脂肪酶LipA活性中心氨基酸残基Ser87(组成活性中心催化三联体...Ser87...His286...Asp264...的氨基酸残基之一) 9 ?范围以内的所有氨基酸残基。(3) 在排除上述活性中心及其临近氨基酸残基的基础上,利用FoldX软件和Rosetta软件对脂肪酶LipA其他位点的氨基酸残基进行虚拟饱和突变,并计算出每个突变体的自由能变化值[14-15]。(4) 以ΔΔG<-0.5 kcal/mol (FoldX软件分析)和ΔΔG<0 kcal/mol (Rosetta软件分析)作为热稳定性提高的突变体的筛选标准,合并上述2个突变电子文库。(5) 利用YASARA软件对上述突变电子文库中的每个突变体的三级结构进行分析,删除结构明显不合理的突变体[7];(6) 剩余的突变体,进一步利用YASARA软件进行分析,筛选出突变位点的氨基酸残基与周围氨基酸残基之间能形成盐桥的突变体,作为本实验的研究对象。
1.3 脂肪酶lipA基因突变体的构建 以质粒pACYC-lipA/lipB为模板,利用表 1中的引物对,PCR扩增全长质粒。PCR扩增程序为:95 ℃ 5 min;95 ℃ 30 s,退火30 s (退火温度参考表 1),72 ℃ 6 min,25个循环;72 ℃ 6 min。PCR扩增产物经电泳鉴定后,加入0.5 μL QuickCut Dpn I限制性内切酶(25 μL PCR反应体系) 37 ℃酶切4 h后,电泳、纯化目的产物,并转化E. coli BL21 (DE3) 感受态细胞。重组转化子经培养后,抽提质粒并送交测序公司进行序列分析。
1.4 脂肪酶lipA基因及其突变体的诱导表达 将测序验证正确的重组转化子转接到5 mL LB培养基(含34 μg/mL的氯霉素)中,37 ℃、220 r/min振荡培养12 h。按2% (V/V)的接种率转接到20 mL LB培养基中(含34 μg/mL的氯霉素,250 mL三角瓶),37 ℃、220 r/min培养至OD600达0.6-0.9时,加入IPTG至终浓度为1 mmol/L。30 ℃、220 r/min培养16 h后,离心收集菌体。用pH 7.4 20 mmol/L Na2HPO4-NaH2PO4缓冲溶液洗涤菌体1次,并重新用8 mL相同的缓冲溶液悬浮菌体。悬浮菌体利用超声破碎仪进行裂解,裂解条件为:处理时间6 min,功率35%,工作2 s,间歇2 s。4 ℃、12000 r/min,离心10 min,收集上清液,作为粗酶液,进行后续分析。
1.5 脂肪酶酶活的测定 脂肪酶酶活的测定采用比色法[15]。具体酶学反应体系及测定方法参照刘艳如等[12]。
1.6 热稳定性提高的脂肪酶突变体的筛选 脂肪酶热稳定性的测定方法参照刘艳如等[12]。比较突变体脂肪酶与野生型脂肪酶在55 ℃处理12 min后的残余酶活,筛选出残余酶活大于野生型脂肪酶的突变体,进行复筛。
1.7 脂肪酶及其突变体T5012和t1/2的测定 T5012和t1/2的测定方法参照Zhao和Arnold[16]。具体实验方法、酶活测定体系等参照刘艳如等[12]。
1.8 分子动力学模拟 利用Gromacs 4.5.6软件在55 ℃条件下对野生型脂肪酶LipA和3个阳性突变体进行分子动力模拟分析,分析其RMSD变化趋势。分子动力学模拟使用的力场为CHARMM27,溶质原子距离盒子边缘的距离设置为0.9 nm。同时,向水溶剂中添加合适的抗衡离子(Na+/Cl-),使模拟系统成电中性。系统能量优化采取最陡下降法(Steepest descents)。经NVT平衡和NPT平衡后,在系统达到温度328 K、压力1.05 bar、密度998.3 kg/m3时,对系统进行时长为1 ns的MD分析[17]。
2 结果和分析 2.1 热稳定性增强且存在盐桥效应的LipA突变体电子文库的构建 利用YASARA软件,对模拟的LipA 3D结构进行分析,以距离Ser87 9 ?为标准,可将Ser87、His286、Asp264、Leu17和Gln88 (前3个氨基酸残基为组成催化三联体的氨基酸残基,后2个氨基酸残基为构成阴离子氧洞的氨基酸残基)等40个氨基酸残基排除在突变范围之外。以ΔΔG<-0.5 kcal/mol为标准,利用FoldX构建的虚拟饱和突变文库含511个突变体;而以ΔΔG<0 kcal/mol为标准,利用Rosetta构建的虚拟饱和突变文库含322个突变体。合并上述2个突变体文库后,构建出1个覆盖率更高、包含711个突变体的电子文库。利用YASARA软件,逐一评估上述711个突变体的3D结构,删除空间结构冲突或空间结构不合理的突变体后,最终获得1个包含341个突变体的微型电子文库(表 2)。对上述341个突变体进一步分析,发现其中27个突变体存在盐桥效应(图 1)。
表 2. 热稳定性脂肪酶LipA突变体电子文库及其对应的自由能变化值 Table 2. The thermostable lipase LipA mutant library designed by computer aid and the corresponding ΔΔG value of every mutant
| Mutation site | △△G/(kcal/mol) | Mutation site | △△G/(kcal/mol) | Mutation site | △△G/(kcal/mol) | Mutation site | △△G/(kcal/mol) |
| G3R | -0.7785 | A128S | -0.6913 | A186T | -0.5100 | V240D | -0.178 |
| G3K | -0.5344 | A128D | -0.4540 | A186V | -0.5580 | V240Q | -0.246 |
| T7V | -0.8362 | V129K | -0.5161 | A186K | -0.6990 | V240K | -0.321 |
| R8K | -0.1200 | A128K | -0.7085 | T192L | -0.5124 | L243T | -0.051 |
| D21Q | -0.1930 | Y129S | -0.8340 | T192N | -0.5048 | L243S | -0.240 |
| D21E | -0.3920 | V129L | -0.7060 | T192H | -0.5344 | T245K | -0.921 |
| A24C | -0.5588 | V129R | -0.9818 | T192P | -1.4413 | T245N | -0.147 |
| A24E | -0.6514 | V129E | -0.2590 | A194L | -0.5913 | L246R | -1.205 |
| A24H | -0.8405 | Y129A | -0.3040 | A194R | -0.6576 | A247L | -0.570 |
| A24G | -1.3081 | Y129T | -0.4570 | A194K | -0.1120 | A247R | -0.871 |
| A24L | -1.3944 | Y129N | -0.4930 | A194N | -0.1400 | A247D | -0.008 |
| A24R | -2.7022 | Y129Q | -0.5260 | A194S | -0.2260 | A247V | -0.115 |
| A24N | -1.3826 | L134R | -0.8425 | A194Q | 0.2890 | A247E | -0.201 |
| A24S | -0.0110 | L134D | -0.5611 | T198R | -0.8394 | A247Q | -0.223 |
| A24D | -0.1850 | S135T | -0.5618 | T198H | -0.5816 | A247I | -0.399 |
| A24K | -1.3030 | S135Q | -0.6608 | T198D | -1.0785 | F249R | -0.517 |
| A24Q | -05320 | S135R | -1.8750 | V199I | -0.9693 | G250N | -0.450 |
| L27I | -0.1160 | S135K | -1.4986 | V199H | -0.6688 | G250A | -0.576 |
| E28P | -2.1582 | S136Q | -1.2996 | V199L | -1.0340 | G250K | -0.767 |
| Q34R | -0.9208 | S136P | -2.4084 | V199R | -2.1772 | G250I | -0.716 |
| Q34K | -0.5994 | S136R | -1.3254 | V199K | -1.3822 | G250T | -0.347 |
| Q38R | -0.8354 | S136K | -1.6446 | H204K | -0.6588 | G250E | -0.507 |
| Q39R | -0.9360 | S136E | -1.1263 | H204I | -0.8610 | M255L | -0.832 |
| N40K | -0.0860 | V138L | -0.9007 | H204L | -1.2558 | Q262E | -1.464 |
| N40R | -0.3750 | V138I | -0.5829 | H204M | -0.5532 | K269E | -1.712 |
| T43R | -0.9446 | S138K | -0.6372 | L206F | -0.7061 | K276D | -0.087 |
| T43L | -1.1647 | A140L | -0.9693 | L206Y | -0.8394 | L278E | -0.112 |
| T43K | -1.1706 | A140K | -0.5391 | A210C | -0.5386 | S279D | -0.533 |
| T43E | -0.0350 | A140D | -1.0960 | A210L | -0.6448 | S281E | -0.659 |
| T43I | -0.3740 | V143L | -0.5951 | A210V | -0.5816 | S281D | -0.109 |
| V44I | -0.6242 | V143I | -0.7605 | A210T | -0.2900 | S281N | -0.124 |
| A47V | -0.9243 | N144R | -0.7610 | L218K | -1.0785 | N285R | -3.381 |
| A67K | -0.2760 | V145L | -0.5312 | L218N | -0.0050 | N285K | -1.936 |
| A67R | -0.6817 | V145I | -0.5654 | L218S | -0.1150 | L294K | -0.645 |
| A67N | -0.0150 | V145R | -0.5241 | L218T | -0.1760 | L294R | -0.517 |
| A67E | -0.0820 | T150R | -0.6940 | S219E | -0.4060 | L294N | -0.378 |
| K70L | -0.6738 | T150C | -0.7912 | S219T | -0.3770 | V296L | -0.715 |
| T71A | -0.5249 | T150K | -0.9236 | S219Q | -0.9693 | V296I | -0.655 |
| T71L | -0.9591 | S152K | -0.6748 | V220I | -2.1772 | V296K | -0.967 |
| T71K | -0.9708 | S153T | -0.4030 | V220T | -0.7750 | A299L | -0.927 |
| T71E | -0.6674 | S153R | -0.6373 | F221D | -1.3700 | A299V | -0.622 |
| T71Q | -0.8760 | S153K | -0.5605 | T224V | -1.0490 | A299I | -0.736 |
| T71I | -1.0157 | S153E | -0.6592 | T224I | -1.328 | A299R | -0.734 |
| T71R | -1.1490 | S153Q | -0.6412 | T224R | -1.308 | A299C | -0.893 |
| T71D | -0.0460 | S153D | -0.6851 | T224K | -1.789 | A299Q | -0.599 |
| V72I | -1.0980 | S153N | -0.0520 | G225A | -0.571 | A299T | -0.047 |
| A74N | -0.0620 | S153V | -0.2740 | G225L | -0.745 | A299K | -1.059 |
| A75Q | -0.0850 | N155D | -0.5002 | G225K | -0.772 | E302P | -0.971 |
| A75I | -0.1030 | T156G | -0.7054 | G225D | -0.876 | E302Q | -0.238 |
| A75K | -0.1090 | T156A | -0.7947 | G225R | -1.012 | V305R | -0.586 |
| A75L | -0.3200 | T156R | -0.9517 | T227K | -0.563 | V305Q | -0.038 |
| A75E | -0.5730 | T156S | -0.6136 | D228K | -1.126 | A306L | -0.555 |
| A78R | -0.9365 | T156C | -0.8422 | T229S | -0.558 | A306S | -0.651 |
| A78S | -0.6248 | T156D | -0.7406 | T229E | -0.919 | A306Q | -0.504 |
| A78K | -0.9667 | T156N | -0.8129 | T229Q | -0.875 | A306E | -0.067 |
| T79P | -0.9351 | T156L | -1.0726 | T229R | -2.095 | A306K | -0.157 |
| T79R | -0.9866 | T156P | -1.0719 | T229K | -1.584 | I308M | -0.642 |
| T79E | -0.7094 | T156K | -1.2328 | T231Q | -0.783 | T310E | -1.226 |
| T79K | -1.0755 | T156E | -1.0154 | T231R | -1.979 | T310I | -1.043 |
| K80R | -0.8256 | T156Q | -1.0131 | L232Q | -0.171 | T310R | -0.556 |
| A97R | -0.8281 | N157D | -0.7379 | L232E | -0.296 | T310K | -0.843 |
| A97K | -1.5113 | L161E | -0.5420 | L232K | -0.458 | T310Q | -0.888 |
| A98I | -0.6848 | Q165K | -0.5024 | L234A | -0.895 | T310N | -0.636 |
| A98L | -1.6157 | T166Q | -1.7472 | L234G | -1.366 | T310S | -0.331 |
| A98R | -2.2462 | T166N | -1.5465 | L234R | -1.456 | A312L | -0.534 |
| A98K | -1.4528 | A170Q | -0.5121 | L234D | -1.209 | A312M | -3.206 |
| A100L | -1.4349 | A170D | -0.5624 | L234N | -0.214 | N313A | -0.632 |
| A100R | -1.3800 | A170E | -0.0770 | L234D | -0.220 | N313K | -0.903 |
| A100K | -1.4495 | A170T | -0.0830 | L234K | -0.012 | N313Q | -0.656 |
| D102R | -0.7704 | A173E | -0.7490 | L234K | -0.309 | N313H | -0.544 |
| D102E | -0.6733 | T174R | -0.9199 | V235G | -0.045 | N313L | -1.870 |
| D102Q | -0.7028 | T174N | -0.0700 | V235A | -0.169 | N313R | -0.136 |
| D102N | -0.5314 | Q177N | -0.0650 | V235E | -0.304 | R314I | -0.043 |
| V104I | -0.7344 | Q177K | -0.3540 | V235T | -0.431 | R314N | -0.249 |
| S117M | -1.3885 | N178Q | -0.9495 | V235S | -0.523 | R314E | -0.322 |
| N125K | -0.0370 | N178L | -1.0417 | V235D | -0.563 | R314A | -0.358 |
| N125T | -0.1110 | N178R | -1.0936 | V235N | -0.631 | R314Q | -0.389 |
| N125L | -0.1610 | N178K | -1.1880 | V235L | -0.811 | R314K | -0.422 |
| N125D | -0.1960 | P180R | -0.8366 | V235I | -0.575 | R314K | -0.518 |
| N125I | -0.2750 | A186E | -0.6911 | V235R | -0.793 | K316R | -0.543 |
| N125Q | -0.7590 | A186S | -0.0020 | V235K | -0.773 | L317R | -0.949 |
| N125E | -1.3540 | A186D | -0.1090 | V235Q | -0.786 | L317K | -0.564 |
| V126L | -0.7786 | A186L | -0.0510 | N239R | -1.611 | L317N | -0.077 |
| V126I | -0.8724 | A186C | -0.2560 | N239K | -2.227 | ||
| A128N | -0.6060 | A186H | -0.2580 | V240E | -0.170 | ||
| A128L | -0.5365 | A1861 | -0.2640 | V240N | -0.141 |
表选项
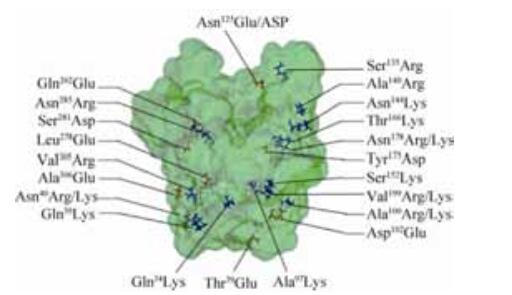 |
| 图 1 在LipA突变体电子文库中存在盐桥效应的突变体 Figure 1 The corresponding mutation sites to form salt bridge in the LipA molecular model. |
| 图选项 |
2.2 突变体的构建及突变效应 上述27个突变体中,LipA-Asn40Arg和LipA-Asn40Lys、LipA-Ala100Arg和LipA-Ala100Lys、LipA-Asn125Asp和LipA-Asn125Glu、LipA-Asn178Arg和LipA-Asn178Lys、LipA-Val199Arg和LipA-Val199Lys为同一位点突变成性质相似的不同氨基酸残基。在第1轮筛选实验中,仅构建了突变体LipA-Asn40Arg、LipA-Ala100Lys、LipA-Asn125Asp、LipA-Asn178Arg和LipA-Val199Lys。上述突变体经验证后,若具有正效应,再构建对应的另外1个突变体(如LipA-Asn125Glu),因此实际仅对23个突变体进行了基因突变的工作(表 1)。最终经测序验证,成功获得20个突变体的重组蛋白。另外3个突变体(Tyr175Asp、Asn178Arg和Asn285Arg),多次实验结果均显示,虽然在突变位点发生了正确的基因突变,但同时在其他位置均存在插入突变。插入突变的引入与上述突变位置附近的核苷酸序列结构特点(如GC含量、回文序列等)有关。后续实验中,PCR扩增体系及条件还有待进一步优化。不同突变体的突变效应如表 3。3个突变体LipA-Asn125Asp、LipA-Asn125Glu和LipA-Gln262Glu较野生型脂肪酶LipA,热稳定性有显著性的提高。55 ℃处理12 min后,突变体LipA-Asn125Asp、LipA-Gln262Glu和LipA-Asn125Glu的残余酶活较野生型LipA分别提高了40.99%、52.3%和57.69%。
表 3. 经热处理后,脂肪酶突变体与野生型脂肪酶残余酶活的比较 Table 3. The difference comparision of the residual activity between LipA mutants and the wild-type LipA
| LipA mutants | Increase rate of residual activity/% | LipA mutants | Increase rate of residual activity/% |
| LipA-Gln34Lys | 3.52±1.14 | LipA-Ala140Arg | 1.25±0.51 |
| LipA-Gln39Lys | -8.899±0.600 | LipA-Asn144Lys | -1.070±1.067 |
| LipA-Asn40Arg | 2.56±0.65 | LipA-Ser152Lys | -8.35±2.15 |
| LipA-Thr79Glu | -1.55±1.22 | LipA-Thr166Lys | -9.12±1.56 |
| LipA-Ala97Lys | -7.83±1.25 | LipA-Val199Lys | -9.60±0.26 |
| LipA-Ala100Lys | -4.23±0.26 | LipA-Gln262Glu | 52.30±2.13 |
| LipA-Asp102Glu | -6.92±1.53 | LipA-Leu278Glu | 0.85±1.53 |
| LipA-Asn125Glu | 57.69±1.43 | LipA-Ser281Asp | -9.80±0.23 |
| LipA-Asn125Asp | 40.99±1.14 | LipA-Val305Arg | -1.210±1.413 |
| LipA-Ser135Lys | -6.39±1.75 | LipA-Ala306Glu | -7.63±2.56 |
表选项
2.3 野生型脂肪酶及正效应脂肪酶突变体T5012的测定 野生型脂肪酶与正效应脂肪酶突变体的T5012测定结果如图 2。野生型脂肪酶LipA的T5012为51 ℃,3个正效应的脂肪酶突变体较野生型脂肪酶的T5012均有一定程度的提高,分别为:55 ℃(LipA-Asn125Asp)、56.5 ℃ (LipA-Asn125Glu)和55.4 ℃(LipA-Gln262Glu)。
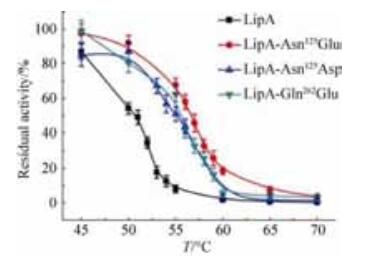 |
| 图 2 野生型脂肪酶及其突变体T5012的测定 Figure 2 The difference in T5012 value between the wild-type LipA and LipA mutants. The T5012 value at each temperature was the average value of the three independent assays and the errors of each T5012 value were presented as the standard deviation. |
| 图选项 |
2.4 野生型脂肪酶及正效应脂肪酶突变体t1/2的测定 野生型脂肪酶与正效应脂肪酶突变体的t1/2测定结果如图 3。野生型脂肪酶LipA的t1/2为5.20 min,突变体LipA-Asn125Asp的t1/2为11.6 min,LipA-Asn125Glu为19.8 min,LipA-Gln262Glu为13.5 min。
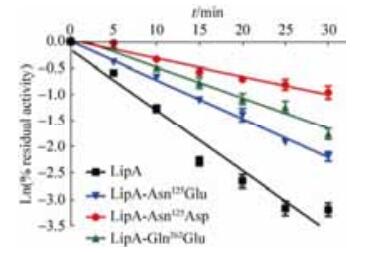 |
| 图 3 野生型脂肪酶及其突变体t1/2的测定 Figure 3 The difference of the t1/2 graph between the wild-type LipA and LipA mutants. The residual activity at each temperature was the average value of the three independent assays and the errors of each ln(% residual activity) value were presented as the standard deviation. |
| 图选项 |
2.5 脂肪酶突变体稳定性提高的分子机制分析 3个正效应脂肪酶突变体在突变位点与周围临近的氨基酸残基之间形成盐桥结构示意图(图 4-A-C)。突变体LipA-Asn125Asp (图 4-A)和突变体LipA-Asn125Glu (图 4-B)的Asp125、Glu125分别与Arg258氨基酸残基之间形成了盐桥。突变体LipA-Gln262Glu的Glu262与Lys269氨基酸残基之间形成了盐桥(图 4-C)。图 4-D显示,3个脂肪酶突变体的RMSD值的波动范围较野生型脂肪酶都有一定的降低,其中LipA-Asn125Glu的RMSD值下降最为明显,分子动力学模拟的结果与前述实验结果基本一致。
 |
| 图 4 脂肪酶突变体热稳定性提高的分子机制 Figure 4 Molecular mechanism for thermostability improvement of three lipase mutants. The schematic model of the salt bridge position in the 3D structural model of LipA mutants was shown in the (A), (B) and (C), respectively. D displayed the overall RMSD values of wied-type LipA and LipA mutants. |
| 图选项 |
3 讨论 热耐受性生物催化剂较中温催化剂而言,可以在较高温度下催化反应,提高催化效率;降低产物被污染的概率;同时,酶制剂具有较好的稳定性,可以提高重复使用批次,从而有效降低生产成本。近些年来,围绕热稳定催化剂资源的挖掘[18]、热稳定蛋白耐热的分子机制[3, 19]、热稳定催化剂突变体的构建[20]及热稳定蛋白突变体的高通量筛选[21]等方面的研究,取得了长足的进展,诞生了一系列新技术新方法。
同定向进化技术中构建突变基因文库一样,电子突变文库的质量直接决定了阳性突变率的高低。Floor等仅从150个突变体的电子文库中筛选到半衰期延长了200多倍的阳性突变体[7];Wijma等从包含64个突变体的电子文库中,筛选到21个热稳定突变体。其中最优热稳定性突变体,表观熔点温度从50 ℃提高到85 ℃,半衰期提高了250多倍[15]。本实验中,虽未对产生的341个突变体的电子文库全部进行实验验证,但验证存在盐桥效应的27个突变体的实验结果表明,仅3个脂肪酶突变体的热稳定性有显著性提高,阳性突变率依然较低。较低的阳性突变率与突变氨基酸的选定范围、筛选突变体自由能ΔΔG的划分标准存在一定的联系。本试验中,我们以9 ?作为活性中心范围的标准。事实上,LipA分子的3D结构具有较深的funnel-like状底物结合部位[22],其活性中心的范围远大于9 ? (如位于活性中心的Ser87的Cα原子与Leu127的Cα原子之间的距离为22.178 ?)。因此,通过扩大活性中心范围的氨基酸残基(本试验中,活性中心的氨基酸残基排除在潜在突变位点之外),可以有效降低突变体文库的容量,一定程度上有助于提高阳性突变率。另外,精确计算突变体自由能,并划定出合理的数值区间,作为筛选、评估突变体的突变效应的标准,也有助于提高阳性突变率。Morloy和Kazlauskas统计了目前报道的各类阳性突变体的ΔΔG自由能变化,发现-ΔΔG值在高于2-3 kcal/mol时,具有良好的阳性突变效应[23]。Christensen和Kepp通过优化FoldX程序并精准计算出ΔΔG的数值,有效提高了预测及评估潜在突变位点的能力[14]。
突变体LipA-Gln262Glu在—Glu262......Lys269—之间形成盐桥,Glu262和Lys269均位于loop环上。通过提高蛋白质柔性区域(flexible region)的刚性(rigidity),实现增强蛋白质的稳定性,已成为构建热稳定蛋白质的常规策略[20, 24-25]。突变体LipA-Asn125Asp和LipA-Asn125Glu形成的盐桥位于LipA分子的α-螺旋α4与α9之间,而α4和α9是构成LipA分子2个盖子结构域(Lid1/Lid2) 的重要二级结构成分(Lid1由α4和α5及其两侧的loop区组成;Lid2由α8、α9、β3和β4及其临近的loop区组成)。脂肪酶的盖子结构域具有高度的柔性和灵活性,在油水界面,盖子结构由闭合状态转变为打开状态,暴露出脂肪酶分子的活性中心,完成脂肪酶的界面激活(Interfacial activation)功能[26-27]。
本实验仅测定了突变体电子文库中存在盐桥效应突变体的突变效应,初步实验结果证实利用生物信息学构建的突变体电子文库具备筛选热稳定性突变体的潜力和价值。进一步深入分析该电子文库中各类突变体提高热稳定性的分子机制,可发现存在多种效应,如二硫键效应(如Ala24Cys/Thr150Cys间可形成二硫键)、脯氨酸效应(如Ser136Pro、Thr156Pro等)、疏水效应、β-转角构象优化等。初步试验结果表明:不同效应叠加产生的突变体,具备更好的热稳定性。如本实验筛选出的具有盐桥效应的突变体继续叠加脯氨酸效应突变产生的叠加突变体,其T5012较野生型LipA提高了10.2 ℃;t1/2延长了18倍,更详细深入的调查仍在进行之中。
References
| [1] | Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme and Microbial Technology, 2006, 39(2): 235-251. DOI:10.1016/j.enzmictec.2005.10.016 |
| [2] | Pleiss J, Fischer M, Peiker M, Thiele C, Schmid RD. Lipase engineering database:understanding and exploiting sequence-structure-function relationships. Journal of Molecular Catalysis B:Enzymes, 2000, 10(5): 491-508. DOI:10.1016/S1381-1177(00)00092-8 |
| [3] | Vieille C, Zeikus GJ. Hyperthermophilic enzymes:sources, uses, and molecular mechanisms for thermostability. Microbiology and Molecular Biology Review, 2001, 65(1): 1-43. DOI:10.1128/MMBR.65.1.1-43.2001 |
| [4] | Royter M, Schmidt M, Elend C, H?benreich H, Sch?fer T, Bornscheuer UT, Antranikian G. Thermostable lipases from the extreme thermophilic anaerobic bacteria Thermoanaerobacter Thermohydrosulfuricus SOL1 and Caldanaerobacter subterraneus subsp. Tengcongensis. Extremophiles, 2009, 13(5): 769-783. DOI:10.1007/s00792-009-0265-z |
| [5] | Reetz MT, Carballeira JD. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nature Protocols, 2007, 2(4): 891-903. DOI:10.1038/nprot.2007.72 |
| [6] | Reetz MT, Kahakeaw D, Lohmer R. Addressing the numbers problem in directed evolution. Chembiochem, 2008, 9(11): 1797-1804. DOI:10.1002/cbic.v9:11 |
| [7] | Floor RJ, Wijma HJ, Colpa DI, Ramos-Silva A, Jekel PA, Szymański W, Feringa BL, Marrink SJ, Janssen DB. Computational library design for increasing Haloalkane dehalogenase stability. ChemBioChem, 2014, 15(11): 1660-1672. DOI:10.1002/cbic.201402128 |
| [8] | Zhou XL, Gao L, Yang GY, Liu DL, Bai AX, Li BC, Deng ZX, Feng Y. Design of hyperthermophilic lipase chimeras by key motif-directed recombination. ChemBioChem, 2015, 16(3): 455-462. DOI:10.1002/cbic.v16.3 |
| [9] | Shu ZY, Lin RF, Jiang H, Zhang YF, Wang MZ, Huang JZ. A rapid and efficient method for directed screening of lipase-producing Burkholderia cepacia complex strains with organic solvent tolerance from rhizosphere. Journal of Bioscience and Bioengineering, 2009, 107(6): 658-661. DOI:10.1016/j.jbiosc.2009.01.011 |
| [10] | Shu ZY, Wu JG, Chen D, Cheng LX, Zheng Y, Chen JP, Huang JZ. Optimization of Burkholderia sp. ZYB002 lipase production for pitch control in thermomechanical pulping (TMP) processes. Holzforschung, 2012, 66(3): 341-348. |
| [11] | Shu ZY, Lin H, Shi SL, Mu XD, Liu YR, Huang JZ. Cell-bound lipases from Burkholderia sp. ZYB002:gene sequence analysis, expression, enzymatic characterization, and 3D structural model. BMC Biotechnology, 2016, 16(1): 38. DOI:10.1186/s12896-016-0269-6 |
| [12] | Liu YR, Qiu LQ, Huang JZ, Zhao BC, Wang ZZ, Zhu XL, Gao YY, Shu ZY. Screening for mutants with thermostable lipase A from Burkholderia sp. ZYB002. Acta Microbiologica Sinica, 2015, 55(6): 748-754. (in Chinese) 刘艳如, 邱黎清, 黄建忠, 赵丙春, 王作镇, 朱晓兰, 高媛媛, 舒正玉. 热稳定性伯克霍尔德菌脂肪酶A突变体的筛选. 微生物学报, 2015, 55(6): 748-754. |
| [13] | Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA-a self-parameterizing force field. Proteins:Structure, Function, and Bioinformatics, 2002, 47(3): 393-402. DOI:10.1002/prot.v47:3 |
| [14] | Christensen NJ, Kepp KP. Accurate stabilities of laccase mutants predicted with a modified FoldX protocol. Journal of Chemical Information and Modeling, 2012, 52(11): 3028-3042. DOI:10.1021/ci300398z |
| [15] | Wijma HJ, Floor RJ, Jekel PA, Baker D, Marrink SJ, Janssen DB. Computationally designed libraries for rapid enzyme stabilization. Protein Engineering, Design and Selection, 2014, 27(2): 49-58. DOI:10.1093/protein/gzt061 |
| [16] | Zhao HM, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Engineering, Design and Selection, 1999, 12(1): 47-53. DOI:10.1093/protein/12.1.47 |
| [17] | Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5:a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics, 2013, 29(7): 845-854. DOI:10.1093/bioinformatics/btt055 |
| [18] | Liszka MJ, Clark ME, Schneider E, Clark DS. Nature versus nurture:developing enzymes that function under extreme conditions. Annual Review of Chemical and Biomolecular Engineering, 2012, 3(1): 77-102. DOI:10.1146/annurev-chembioeng-061010-114239 |
| [19] | Li WF, Zhou XX, Lu P. Structural features of thermozymes. Biotechnology Advances, 2005, 23(4): 271-281. DOI:10.1016/j.biotechadv.2005.01.002 |
| [20] | Yu HR, Huang H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnology Advances, 2014, 32(2): 308-315. DOI:10.1016/j.biotechadv.2013.10.012 |
| [21] | Bommarius AS, Broering JM, Chaparro-Riggers JF, Polizzi KM. High-throughput screening for enhanced protein stability. Current Opinion in Biotechnology, 2006, 17(6): 606-610. DOI:10.1016/j.copbio.2006.10.001 |
| [22] | Kim KK, Song HK, Shin DH, Hwang KY, Suh SW. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure, 1997, 5(2): 173-185. DOI:10.1016/S0969-2126(97)00177-9 |
| [23] | Morley KL, Kazlauskas RJ. Improving enzyme properties:when are closer mutations better?. Trends in Biotechnology, 2005, 23(5): 231-237. DOI:10.1016/j.tibtech.2005.03.005 |
| [24] | Mamonova TB, Glyakina AV, Galzitskaya OV, Kurnikova MG. Stability and rigidity/flexibility-two sides of the same coin?. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2013, 1834(5): 854-866. DOI:10.1016/j.bbapap.2013.02.011 |
| [25] | Rathi PC, Fulton A, Jaeger KE, Gohlke H. Application of rigidity theory to the thermostabilization of lipase A from Bacillus subtilis. PLoS Computational Biology, 2016, 12(3): e1004754. DOI:10.1371/journal.pcbi.1004754 |
| [26] | Cheng M, Angkawidjaja C, Koga Y, Kanaya S. Requirement of lid2 for interfacial activation of a family I. 3 lipase with unique two lid structures. The FEBS Journal, 2012, 279(19): 3727-3737. DOI:10.1111/j.1742-4658.2012.08734.x |
| [27] | Barbe S, Lafaquière V, Guieysse D, Monsan P, Remaud-Siméon M, André I. Insights into lid movements of Burkholderia cepacia lipase inferred from molecular dynamics simulations. Proteins:Structure, Function, and Bioinformatics, 2009, 77(3): 509-523. DOI:10.1002/prot.v77:3 |
