徐玲霞, 王筱兰

 , 杨慧林, 杨林, 施文杰, 谢雅雯, 林姗
, 杨慧林, 杨林, 施文杰, 谢雅雯, 林姗 江西师范大学生命科学学院,江西省亚热带植物资源保护与利用重点实验室,江西 南昌 330022
收稿日期:2015-11-26;修回日期:2016-02-08;网络出版日期:2016-02-19
基金项目:江西省科技厅支撑计划项目(20142BBG70016);江西省自然科学基金(20142BAB214008, 20151BAB204003);江西省亚热带植物资源保护与利用重点实验室开放基金(YRD201405)
*通信作者:王筱兰, Tel: +86-791-88120391; E-mail: xlwang08@aliyun.com
摘要: [目的]新金分枝杆菌(Mycobacterium neoaurum)MN4是1株经诱变育种获得的高产雄烯二酮,并且能够耐受高浓度底物植物甾醇的突变菌株。为深入研究MN4菌株耐底物的机制及雄烯二酮的生物合成途径,有必要解析MN4菌株的全基因组序列信息。[方法]本研究采用高通量测序技术对MN4进行全基因组测序,然后使用相关软件对测序数据进行基因组装、基因预测与功能注释、COG聚类分析以及次级代谢产物合成基因簇预测等。[结果]新金分枝杆菌MN4基因组装获得33个Contigs,整个基因组大小为5.39 Mb,GC含量为66.9%,编码4920个蛋白基因,序列提交至GenBank数据库,登录号为JXYZ00000000。[结论]本研究首次报道了1株高产雄烯二酮菌株MN4的全基因组序列,分析了基因组的基本特征,初步解析了该菌株降解植物甾醇生产雄烯二酮的关键基因,将为MN4的功能基因组学研究及相关次级代谢产物的生物合成途径与异源表达研究提供基础。
关键词: 雄烯二酮 新金分枝杆菌 甾醇 全基因组测序 生物合成基因簇
Whole-genome sequencing and analysis of a adrostenedione yielding strains Mycobacterium neoaurum MN4
Lingxia Xu, Xiaolan Wang

 , Huilin Yang, Lin Yang, Wenjie Shi, Yawen Xie, Shan Lin
, Huilin Yang, Lin Yang, Wenjie Shi, Yawen Xie, Shan Lin Key Laboratory of Protection and Utilization of Subtropic Plant Resources, College of Life Science, Jiangxi Normal University, Nanchang 330022, Jiangxi Province, China
Received 26 November 2015; Revised 08 February 2016; Published online 19 February 2016
*Corresponding author: Xiaolan Wang.Tel: +86-791-88120391; E-mail: xlwang08@aliyun.com
Supported by the Science and Technology Support Program of Jiangxi Province (20142BBG70016), by the Natural Science Foundation of Jiangxi Province (20142BAB214008, 20151BAB204003) and by the Development Foundation of Key Laboratory of Protection and Utilization of Subtropic Plant Resources (YRD201405)
Abstract: [Objective]Mycobaterium neoaurum MN4 is a substrate-resistant mutant strain with high-yield androstenedione. In order to further study MN4 strain substrate-resistant mechanism and androstenedione biosynthetic pathway, it is necessary to decipher the MN4 strain genome.[Methods]The genome was sequenced using high-throughput sequencing technology, and analyzed using relevant software for genome assembly, gene prediction and functional annotation, COG cluster analysis and secondary metabolite biosynthesis gene clusters prediction.[Results]The whole genome is assembled into 33 contigs, and the genome size is 5.39 Mb, GC content of 66.9% with encoding 4920 protein genes. The genome sequence was deposited in the GenBank database under the accession number JXYZ00000000.[Conclusion]This study is the first report of androstenedione producing strain Mycobacterium neoaurum MN4 genome sequence, and provides a theoretical basis for further heterologous expression of secondary metabolites on Mycobacterium neoaurum MN4.
Key words: androstenedione Mycobaterium neoaurum steroids whole-genome sequencing biosynthesis gene cluster
甾醇类化合物(Steroids)又称类固醇,是在生物体组织内广泛存在的化合物,从结构上来看,是环戊烷多氢菲核的化合物的一类。截止目前,所经授权上市的甾体类药物共有400多种,全球总产值已达到400亿美元,而且每年呈递增的趋势,甾类药物的市场需求多年来名列前茅,仅次于抗生素[1]。甾类激素药物在临床中应用非常的广泛:改善人类生育能力及抗骨质疏松,调节血压[2-4];具有显著的抗肿瘤、抗炎抗过敏、抗菌抗病毒等效果,而且对于一些由激素水平紊乱所造成的恶疾具有一定的治愈效果[5-7];可作为抗肥胖药物的活性成分,以及预防HIV的传染和艾滋病的治疗等。
雄烯二酮(Androst-4-ene-3, 17-dione, AD)是生产甾体激素药物不可替代的关键中间体,几乎所有的甾体激素药物都是以其作为起始原料进行生产的[8-9]。20世纪60年代中期,美国生物学家Sih等[10]首先发现微生物能够切除饱和甾醇的侧链得到AD和ADD。20世纪70年代初,日本的马启利用微生物降解胆甾醇生产ADD也获得了成功[11]。如今,AD被一些发达国家用于开发和生产新的医药主品,如福美司坦和类固醇类激素免疫抗原等。目前在全世界范围内,AD做原材料生产的医药品种不断增加,为世界上的广大患者带来了福音[12]。
放线菌属革兰氏阳性、好养型微生物,多生活在土壤环境中,可代谢产生或合成多种次级代谢产物,例如链霉菌大约可以合成2/3的药用抗生素[13],假单胞菌能够有效代谢长链石油烃类[14],分枝杆菌与红球菌等则可以特异性的降解多环甾类物质[15-16]。
本实验室前期通过紫外、硫酸二乙酯复合诱变获得了1株高产雄烯二酮,并且能够耐受高浓度底物植物甾醇的新金分枝杆菌(Mycobacterium neoaurum)MN4[17],并保藏于中国典型培养物保藏中心(CCTCC NO: M 2014322)。为深入研究MN4菌株耐底物的机制及雄烯二酮的生物合成途径,本研究采用高通量测序技术对MN4菌株进行全基因组测序,并对该菌株的次级代谢产物合成基因簇进行预测,相关研究结果将为MN4的功能基因组学研究及相关次级代谢产物的生物合成途径与异源表达研究提供基础。
1 材料和方法 1.1 主要试剂和仪器 胰蛋白胨、酵母提取物购自于英国OXOID公司,葡萄糖、Tris、EDTA、琼脂粉等均为国产分析纯。上海卢湘仪TGL-20M离心机、BIO-RAD ChemiDoc XRS凝胶成像系统、Illumina HiSeq 2500高通量测序仪。
1.2 分枝杆菌的培养与培养基 新金分枝杆菌MN4由本实验室通过诱变筛选获得,并保藏于中国典型培养物保藏中心(CCTCC NO: M 2014322)。斜面培养基(g/L):酵母膏3,蛋白胨5,葡萄糖20,琼脂粉20,pH 7.0,115℃高压灭菌20 min。液体培养基(g/L):胰蛋白胨10,酵母提取物5,NaCl 20,pH 7.0,115℃高压灭菌20 min。
1.3 分枝杆菌的培养与基因组DNA提取 MN4菌株接种至斜面培养基于31℃活化培养6 d,然后转接至摇瓶液体培养基于31℃、200 r/min培养4 d,然后抽滤收集菌体用于后续基因组DNA提取。采用液氮研磨的方法裂解细胞壁,然后采用OMEGA细菌基因组DNA提取试剂盒抽提总DNA,操作步骤见试剂盒说明书。
1.4 基因组的测序、组装及注释 基因组DNA提取后,在浓度和纯度达到测序要求后进行全基因组测序,后采用Illumina Hiseq 2500测序平台完成基因组测序,采用拼接软Velevt1.2.10[18]进行序列组装,通过优化参数Kmer值获得最好的组装结果。基因预测采用Glimmer 3.02[19]软件进行开放阅读框(ORF)预测,将所有预测蛋白序列与COG(直系同源基因数据库)、NR(非冗余蛋白数据库)、Swiss-Prot数据库[20]、Interpro数据库[21]、KEGG(Kyoto Encyclopedia of Genes and Genomes,京都基因和基因组百科全书)[22]和Gene Ontology (GO)数据库[23]进行BLASTp (E-value≤1e-5)对比,完成蛋白序列功能注释。采用tRNAscan-SE[24]和RNAmmer[25]软件分别进行tRNA和rRNA预测。
1.5 次级代谢产物合成基因簇分析 次级代谢产物的编码基因通常在基因组中成簇存在,本研究采用antiSMASH[26-27]软件对MN4菌株中次级代谢产物合成基因簇进行分析,并预测可能合成的化合物。
2 结果和分析 2.1 比较基因组学分析
2.1.1 Mycobacterium neoaurumMN4基因组的基本特征 目前,已报道有4株Mycobacterium neoaurum完成全基因组测序,分别为Mycobacterium neoaurum MN4、VKMAc-1815D、ATCC 25795和DSM 44074。通过比较研究4株Mycobacterium neoaurum和1株Mycobacterium tuberculosis的基因组基本特征发现:4株新金分枝杆菌在基因组大小、GC含量、CDS数目、CDS平均长度和RNA基因数等方面非常相似(表 1),但与Mycobacterium tuberculosis差异较大,表明MN4符合新金分枝杆菌基因组的基本特征。 表 1. 分枝杆菌基因组基本特征比较分析 Table 1. General features of several sequenced Mycobacterium chromosomes
| Features | Length size/bp | GC content/% | CDS (No.) | Contigs (No.) | Average CDS size/bp | Coding percent/% | tRNA number | Pseudogene (No.) | GenBank No. |
| MN4 | 5390529 | 66.9 | 4920 | 33 | 992 | 90.6 | 46 | 79 | JXYZ00000000 |
| VKMAc-1815 | 5421267 | 66.9 | 4948 | 1 | 996 | 90.9 | 46 | 25 | CP006936 |
| ATCC 25795 | 5468381 | 66.7 | 5059 | 42 | 990 | 91.6 | 46 | 22 | JMDW00000000 |
| DSM 440744 | 5536033 | 66.6 | 5030 | 45 | 978 | 88.9 | 46 | 82 | CCDR000000000 |
| H37RV | 4411532 | 65.6 | 3906 | 1 | 1013 | 89.70 | 45 | 30 | AL123456 |
表选项
MN4、VKMAc-1815D、ATCC 25795和DSM 44074都能够进行降解植物甾醇侧链产生AD和ADD。与其它3株新金分枝杆菌不同,本研究的MN4菌株是实验室前期通过紫外、硫酸二乙酯复合诱变获得了1株高产雄烯二酮(AD),并且能够耐受高浓度底物植物甾醇的突变株,其副产物ADD比较少[17]。Mycobacterium neoaurum ATCC 25795是通过基因工程改造提高AD产率的工程菌株[28]。
2.1.2 Mycobacterium neoaurum MN4编码蛋白质的COG聚类分析 基于Glimmer3.0软件,Mycobacterium neoaurum MN4共预测得到4920个蛋白基因,然后将预测得到的所有蛋白质序列与COG数据库进行BLASTp比对分析(E-value≤1e-5),共计4769个蛋白基因成功获得COG功能注释(图 1)。从图中可以看出,COG聚类主要集中于一般功能预测(General function prediction only)、能量产生及传递(Energy production and conversion)、转录(Transcription)、氨基酸代谢及转运(Amino acid transport and metabolism)、脂类转运及代谢(Lipid transport and metabolism)、次级代谢产物合成(Secondary metabolites biosynthesis, transport and catabolism),分别占14.7%、9.7%、8.9%、8.9%、7.5%、7.2%。值得注意的是,次级代谢产物合成相关基因达345个,如此多的次级代谢产物合成基因极有可能是该菌株能够产生多种次级代谢产物的原因。
 |
| 图 1. Mycobacterium neoaurum MN4蛋白质COG聚类分析 Figure 1. COG cluster analysis of Mycobacterium neoaurum MN4. |
| 图选项 |
2.2 Mycobacterium neoaurum MN4次级代谢产物合成基因簇分析 次级代谢产物的编码基因通常在基因组中成簇存在,编码具有多种功能的复合酶。其中研究的最为清楚是PKS/NRPS基因簇。本研究采用antiSMASH软件对MN4菌株中所有的次级代谢产物合成基因簇进行分析,预测共得到17个基因簇,其中PKS/NRPS基因簇有7个,约占总基因簇数量的41.1%(表 2)。同时我们对部分次级代谢产物的核心结构进行了预测(图 2)。
表 2. Mycobacterium neoaurum MN4次级代谢产物合成基因簇预测 Table 2. Gene clusters involved in synthesis of secondary metabolites in Mycobacterium neoaurum MN4
| ID | Contig No. | Position/nt | Gene amount | Gene cluster type |
| 1 | Contig01 | 159507-180451 | 22 | terpene |
| 2 | Contig01 | 175699-233113 | 41 | nrps |
| 3 | Contig01 | 384839-395636 | 13 | bacteriocin |
| 4 | Contig01 | 429422-472904 | 40 | other |
| 5 | Contig01 | 495629-543243 | 36 | Nrps-t1pks |
| 6 | Contig04 | 113694-124188 | 12 | ectoine |
| 7 | Contig04 | 254297-306672 | 40 | Nrps-t1pks |
| 8 | Contig07 | 80710-125026 | 41 | other |
| 9 | Contig10 | 39439-104907 | 50 | Nrps-t1pks |
| 10 | Contig11 | 57416-116582 | 48 | Nrps-t1pks |
| 11 | Contig13 | 126361-142490 | 17 | terpene |
| 12 | Contig15 | 88473-118128 | 31 | T1pks |
| 13 | Contig17 | 13586-59018 | 31 | T1pks |
| 14 | Contig20 | 32834-66829 | 36 | other |
| 15 | Contig21 | 34844-63419 | 32 | other |
| 16 | Contig23 | 22162-60458 | 29 | other |
| 17 | Contig29 | 1-8702 | 10 | Lassopetide |
| *T1pks: Type I polyketide synthases; nrps: Non-ribosomal peptide; ectoine: 1, 4, 5, 6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. | ||||
表选项
 |
| 图 2. Mycobacterium neoaurum MN4次级代谢产物预测核心结构示意图(部分) Figure 2. Predicted core structure involved in synthesis of secondary metabolites in MN4 (Partial). |
| 图选项 |
antiSMASH软件预测次级代谢产物合成基因簇,其原理是基于BLAST同源比对的方法。我们通过该软件预测到17个可能的次级代谢产物生物合成基因簇,表明MN4菌株具有合成这些代谢产物的潜力和基因元件,具体能否合成这些次级代谢产物还与其培养条件等因素有关。由于新金分枝杆菌的次级代谢产物文献报道较少,未能就预测的次级代谢产物与已知分离得到的次级代谢产物对应上,因此我们对其它微生物中能够合成这些次级代谢产物的菌株及相关文献进行了汇总(表 3)。
表 3. MN4菌株预测合成的次级代谢产物产生菌的文献报道 Table 3. Literature reports of predicted secondary metabolites in MN4 strains
| Predict product | Sources of species | MN4 cluster | References |
| isorenieratene | Green sulfur bacteria | cluster 1 | Maresca JA, et al[29] |
| ectione | Mycobacterium smegmatis | cluster 6 | Ofer N, et al[30] |
| glycopetidolipid | Mycobacterium avium | cluster 7 | Belisle JT, et al[31] |
| galbonolides | Streptomyces sp. LZ35 | cluster 8 | Liu C, et al[32] |
| mycobactin | Mycobacterium tuberculosis | cluster 9 | Reddy PV, et al[33] |
| herboxidene | Streptomyces sp. A7847 | cluster 14 | Isaac BG, et al[34] |
| BE-7585A | Amycolatopsis orientalis | cluster 16 | Yu X, et al[35] |
表选项
2.3 Mycobacterium neoaurum MN4降解甾体基因簇分析 由于已经进行测序的新金分枝杆菌只有4株,而Mycobacterium neoaurum NwIB-01没有进行全基因组测序,故将MN4与同源性最高的VKM AC-1815D (GenBank No. NC_023036)进行比对分析,从而确定了MN4菌株中甾体降解基因簇相关信息(图 3)。
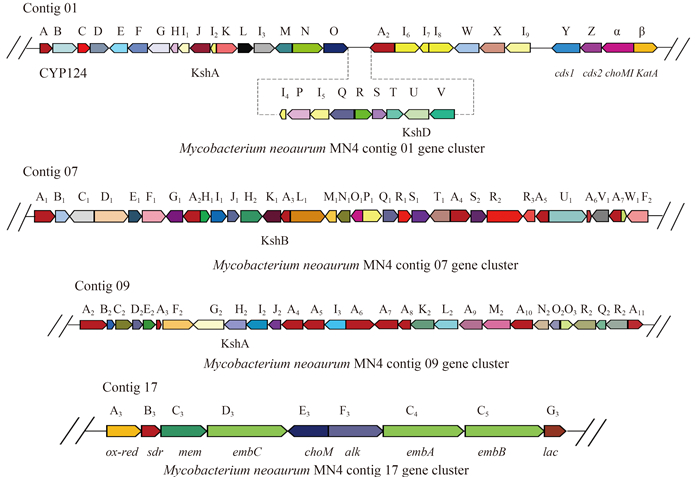 |
| 图 3. Mycobacterium neoaurum MN4甾体降解关键基因簇 Figure 3. Steroidal degradation of key gene cluster of Mycobacterium neoaurum MN4. |
| 图选项 |
华东理工大学魏东芝教授团队在菌株NwIB-01中发现了多个关键基因kstR (GenBank No. GU363925)、kshA (GenBank No. GQ476982)、choM1,KatA (GenBank No. JQ303323)、choM2,embA (GenBank No. JQ303324)、ksdD (GenBank No. GQ228843)等与菌株降解甾体类物质有关[28]。通过BLAST比对分析发现在MN4也存在这些基因,包括关键基因choM1 (GenBank No. KJQ52133);choM2 (GenBank No. KJQ48263);ksdD (GenBank No. KJQ52289);kshA (GenBank No. KJQ51625);kshB (GenBank No. KJQ49581)。
choM1基因(位于contig 01)和choM2基因(位于contig 17)是M. neoaurum MN4降解植物甾醇中的胆固醇氧化酶(CHO)的作用,经过CHO催化后,甾醇氧化为具有4-烯-3-酮的的甾酮,由此开始ksdD基因(位于contig 01)的脱氢作用A环的C1(2)将形成AD以及将AD转化成ADD,然后kshB(位于contig 07)和kshA(位于contig 09)所组成的kSH基因将降解过程中的AD和ADD转化成9-OH AD和9-OH ADD。3-甾酮-9α-羟基化酶(3-ketosteroid-9-alpha-hydroxylase KSH)由末端加氧酶(KshB)与(KshA)双组份构成。KSH催化的羟基反应,开始于NADH的氧化,自由电子通过还原酶KshB辅酶FAD与铁硫还原中心的传递,过渡到氧化酶KshA的Rieske结构域,最终在KshA活性中心Fe2+处,与氧分子结合,完成甾体C9位的加氧反应[36]。M. neoaurum MN4降解植物甾醇的具体途径见图 4。
 |
| 图 4. MN4降解植物甾醇途径[28] Figure 4. Phytosterols degradation pathway of M. neoaurum MN4[28]. CHO: cholesterol oxidase, KSH: 3-ketosteroid-9-alpha-hydroxylase, ksdD: 3-ketosteroid-delta-1-dehydrogenase. |
| 图选项 |
3 讨论 目前能够进行降解植物甾醇侧链制备AD和ADD的菌株主要有分枝杆菌、节杆菌和简单包杆菌等[37]。如今,以甾醇为原料的微生物转化工业体系中,对工业用菌的选育一直是甾体转化中的关键问题。Mycobacterium neoaurum MN4是一以MN2为出发菌株采用紫外与硫酸二乙酯复合诱变处理的突变株,其AD生成率较出发菌株提高了15%,出发菌株MN2转化产物中AD与ADD的比例为13:1,而在突变菌株MN4的转化产物中,AD与ADD的比例为20:1[17]。因此,对MN4进行全基因组测序有助于从分子水平上阐明其高产AD的机制。
微生物降解甾体属有氧代谢,研究认为甾体类物质的代谢起始于甾核的开环与侧链的末端氧化。甾核的开环反应始于胆固醇氧化酶(ChO)的作用,其次是3-甾酮-△1-脱氢和3-甾酮-9α-羟基化反应。经胆固醇氧化酶(ChO)的催化后,甾醇氧化为具有4-烯-3-酮的甾酮,然后开始甾体环的降解代谢,再经3-甾酮-3-甾酮-△1-脱氢酶(KsdD)与3-甾酮-9α-羟基化酶(KsH)的作用形成甾药前体,如ADD和9-OHAD等。KstD是形成ADD等甾药中间体的关键药物酶,但同时也是微生物累积AD和9-OHAD的不利因素。在菌株MN4中只发现了一处KsdD编码序列,从而降低了AD转化为ADD的活力,这或许是导致MN4高产AD的原因。
本研究通过全基因组测序方法,获得了MN4的全基因组序列,预测得到4920个蛋白基因,预测了菌株降解甾醇基因簇。通过比较分析该菌株实际有1处ksdD (GenBank No. KJQ52289)编码序列,2处不同的kshA (GenBank No. KJQ51625, KJQ49194)编码序列及1处kshB (GenBank No. KJQ49581)编码序列。故可以通过基因工程的方法对该菌株中关键基因酶3甾酮-△1脱氢酶(KsdD)进行敲除或强化操作,以筛选出具有高活性或低活性3甾酮-△1脱氢酶(KsdD)的突变株,使产物中AD或ADD的产量占绝对优势,或者将3-甾酮-9α-羟基脱氢酶(KsH)基因进行敲除或强化操作以筛选出具有高活性或低活性3-甾酮-9α-羟基脱氢酶(KsH)的突变株,使产物中9-OHAD或9-OHADD的产量占绝对优势。本研究的相关结果将为Mycobacterium neoaurum MN4的功能基因组学研究及相关次级代谢产物合成途径及异源表达研究提供帮助。
参考文献
| [1] | Tong WY, Dong X. Microbial biotransformation: recent developments on steroid drugs.Recent Patents on Biotechnology, 2009, 3(2): 141–153. |
| [2] | Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, Visvader JE. Control of mammary stem cell function by steroid hormone signalling.Nature, 2010, 465(7299): 798–802. |
| [3] | Callewaert F, Boonen S, Vanderschueren D. Sex steroids and the male skeleton: a tale of two hormones.Trends in Endocrinology and Metabolism, 2010, 21(2): 89–95. |
| [4] | Rubtsov AV, Rubtsova K, Kappler JW, Marrack P. Genetic and hormonal factors in female-biased autoimmunity.Autoimmun Reviews, 2010, 9(7): 494–498. |
| [5] | B?ckstr?m T, Haage D, L?fgren M, Johansson IM, Str?mberg J, Nyberg S, Andréen L, Ossewaardge L, van Wingen GA, Turkmen S, Bengtsson SK. Paradoxical effects of GABA-A modulators may explain sex steroid induced negative mood symptoms in some persons.Neuroscience, 2011, 191: 46–54. |
| [6] | Douglas M. Neurology of endocrine disease.Clinical Medicine, 2010, 10(4): 387–390. |
| [7] | Garcia-Segura LM, Balthazart J. Steroids and neuroprotection: new advances.Frontiers in Neuroendocrinology, 2009, 30(2): 4–5. |
| [8] | Conner AH, Nagaoka M, Rowe JW, Perlman D. Microbial conversion of tall oil sterols to C19 steroids.Applied and Environmental Microbiology, 1976, 32(2): 310–311. |
| [9] | He XJ, Liu B, Wang GH, Wang XL, Su LN, Qu GX, Yao XS. Microbial metabolism of methyl protodioscin by Aspergillus niger culture-a new androstenedione producing way from steroid.The Journal of Steroid Biochemistry and Molecular Biology, 2006, 100(1/3): 87–94. |
| [10] | Sih CJ, Wang KC. A new route to estrone from sterols.Journal of the American Chemical Society, 1965, 87(6): 1387–1388. |
| [11] | Zhang XY, Ruan H, He GQ. Advances in micobial transformation of phytosterols to androstenedione.Journal of Chinese Institute of Food Science and Technology, 2012, 12(7): 162–170(in Chinese).张小燕, 阮晖, 何国庆. 微生物降解植物甾醇侧链生产雄甾烯二酮的研究进展.中国食品学报, 2012, 12(7): 162–170. |
| [12] | Yang Y, Jiang ST. Advance in degrading side chain of sterols of getting androst-4-ene-3, 17-dione by microorganism.Microbiology China, 2006, 36(6): 142–145(in Chinese).杨英, 姜绍通. 微生物降解甾醇侧链转化雄甾-4-烯-3, 17-二酮的研究进展.微生物学通报, 2006, 36(6): 142–145. |
| [13] | Hopwood DA. Streptomyces in nature and medicine: the antibiotic makers.Journal of the History of Medicine and Allied Sciences, 2008, 63(2): 273–274. |
| [14] | Ron EZ, Rosenberg E. Biosurfactants and oil bioremediation.Current Opinion in Biotechnology, 2002, 13(3): 249–252. |
| [15] | Kieslich K. Microbial side-chain degradation of sterols.Journal of Basic Microbiology, 1985, 25(7): 461–474. |
| [16] | Marsheck WJ, Kraychy S, Muir RD. Microbial degradation of sterols.Applied Microbiology, 1972, 23(1): 72–77. |
| [17] | Sun WJ, Wang XL. Screening of substrate-resistant mutant strains producing androst-4-ene-3, 17-dione and optimization of biotransformation medium.Food Science, 2014, 35(19): 158–163(in Chinese).孙婉菊, 王筱兰. 雄烯二酮耐底物突变株的筛选及生物转化培养基优化.食品科学, 2014, 35(19): 158–163. |
| [18] | Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs.Genome Research, 2008, 18(5): 821–829. |
| [19] | Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer.Bioinformatics, 2007, 23(6): 673–679. |
| [20] | Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003.Nucleic Acids Research, 2003, 31(1): 365–370. |
| [21] | Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischman W, Gouzy J, Giffith-Jones S, Haft D, Hermiakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJA. InterPro: an integrated documentation resource for protein families, domains and functional sites.Briefings in Bioinformatics, 2002, 3(3): 225–235. |
| [22] | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes.Nucleic Acids Research, 2000, 28(1): 27–30. |
| [23] | Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck k, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The gene ontology (GO) database and informatics resource.Nucleic Acids Research, 2004, 32(Suppl 1): D258–D61. |
| [24] | Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence.Nucleic Acids Research, 1997, 25(5): 955–964. |
| [25] | Lagesen K, Hallin P, R?dland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes.Nucleic Acids Research, 2007, 35(9): 3100–3108. |
| [26] | Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers.Nucleic Acids Research, 2013, 41(W1): W204–W2012. |
| [27] | Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences.Nucleic Acids Research, 2011, 39(Suppl 2): W339–W346. |
| [28] | Yao K, Xu LQ, Wang FQ, Wei DZ. Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3, 17-dione through the catabolism of sterols.Metabolic Engineering, 2014, 24: 181–191. |
| [29] | Maresca JA, Romberger SP, Bryant DA. Isorenieratene biosynthesis in green sulfur bacteria requires the cooperative actions of two carotenoid cyclases.Journal of Bacteriology, 2008, 190(19): 6384–6391. |
| [30] | Ofer N, Wishkautzan M, Meijler M, Wang Y, Speer A, Niederweis M, Gur E. Ectoine biosynthesis in Mycobacterium smegmatis.Applied and Environmental Microbiology, 2012, 78(20): 7483–7486. |
| [31] | Belisle JT, McNeil MR, Chatterjee D, Inamine JM, Brennan PJ. Expression of the core lipopeptide of the glycopeptidolipid surface antigens in rough mutants of Mycobacterium avium.The Journal of Biological Chemistry, 1993, 268(14): 10510–10516. |
| [32] | Liu C, Zhu J, Li YY, Zhang JL, Lu CH, Wang HX, Shen YM. In vitro reconstitution of a PKS pathway for the biosynthesis of galbonolides in Streptomyces sp LZ35.ChemBioChem, 2015, 16(6): 998–1007. |
| [33] | Reddy PV, Puri RV, Chauhan P, Kar R, Rohilla A, Khera A, Tyagi AK. Disruption of mycobactin biosynthesis leads to attenuation of Mycobacterium tuberculosis for growth and virulence.The Journal of Infectious Diseases, 2013, 208(8): 1255–1265. |
| [34] | Isaac BG, Ayer SW, Elliott RC, Stonard RJ. Herboxidiene: a potent phytotoxic polyketide from Streptomyces sp. A7847.Journal of Organic Chemistry, 2002, 57(26): 7220–7226. |
| [35] | Yu X, Mets?-ketel? M, Tsai SC, Liu HW, Rohr J. Roles of the key enzymes involved in the angucycline core formation in the BE-7585A biosynthesis.Planta Medica, 2014, 80(10): 823. |
| [36] | van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1.Molecular Microbiology, 2002, 45(4): 1007–1018. |
| [37] | Sripalakit P, Wichai U, Saraphanchotiwitthaya A. Biotransformation of various natural sterols to androstenones by Mycobacterium sp. and some steroid-converting, microbial strains.Journal of Molecular Catalysis B:Enzymatic, 2006, 41(1/2): 49–54. |
