鲍文英, 江经纬, 周云, 吴玉峰, 梁志清


南京农业大学生物信息学中心, 江苏 南京 210095
收稿日期: 2015-07-06; 修回日期: 2015-09-20; 网络出版日期: 2015-10-19
通讯作者: 梁志清,Tel:+86-25-84399042;Fax:+86-25-84399047;E-mail:fcleung.njau@gmail.com
摘要: [目的] 筛选和鉴定有木质纤维素降解能力的1株细菌,测定其相关酶活力并进行全基因组分析,为构建木质纤维素降解工程菌提供依据。[方法] 采用3种木质素类似物(天青-B;酚红;愈创木酚)的脱色/染色法,从腐木和被枝叶覆盖的土壤中分离和筛选出1株具有较强木质纤维素降解能力的细菌。通过16S rRNA基因和全基因组序列分析对该菌进行种属鉴定。使用紫外分光光度法测定其锰过氧化物酶(MnP)、漆酶(Lac)、羧甲基纤维素酶(CMCase)以及滤纸酶(FPA)活力,了解该菌相关酶活力大小在一定时间内的变化趋势。使用Illumina Miseq和454 GS Junior测序平台获取该菌的全基因组序列,将其全基因组序列经过注释的基因蛋白质序列提交COG和KEGG数据库进行BLASTp比对分析,确定该菌潜在的重要酶类和代谢途径,并对部分注释基因进行定量RT-PCR验证。[结果] 筛选得到1株优势菌株S12,该菌经鉴定后命名为解鸟氨酸拉乌尔菌(Raoultella ornithinolytica)。在液体CMC-Na培养基中发酵28 h,菌体生长达到稳定期,纤维素降解相关酶活力也在此时达到峰值。生物信息学分析结果表明,菌株S12具有木质素降解通路中重要酶类的编码基因,如过氧化物酶、Fe-Mn型超氧化物歧化酶、邻苯二酚1,2-双加氧酶和原儿茶酸-3,4-双加氧酶等,这些基因在以碱性木质素为碳源的培养条件下表达量不同程度地高于以葡萄糖为碳源的培养条件。另外,菌株S12具备完整的纤维素降解和乙醇生成通路。[结论] 本研究首次揭示了Raoultella ornithinolytica S12具备有效的木质纤维素降解性能,这对于推动木质纤维素应用产业的发展具有重要意义。
关键词: 木质纤维素降解菌筛选拉乌尔属全基因组代谢通路
Screening and genomic analysis of a lignocellulose degrading bacterium
Wenying Bao, Jingwei Jiang, Yun Zhou, Yufeng Wu, Frederick Chi-Ching Leung


Bioinformatics Centre, Nanjing Agricultural University, Nanjing 210095, Jiangsu Province, China
Received: 6 July 2015; Revised: 20 September 2015; Published online: 19 October 2015
Corresponding author. Tel: +86-25-84399042; Fax: +86-25-84399047; E-mail: fcleung.njau@gmail.com
Abstract:[Objective] This study is aimed to screen and identify a bacterium with the ability to degrade lignocellulose, to perform its genomic analysis, and to determine its related enzymatic activities. [Methods] Using a bleaching/dyeing method with three kinds of lignin analogues (Azure-B; Phenol red; Guaiacol), we separated and screened a bacterium strain, with a strong ability to degrade lignocellulose, from soil enriched by decaying wood and leaves. We identified the species of this bacterium according to its 16S rRNA gene and core gene sequence analysis. In order to understand the trend of enzymatic activities within a certain period, we used ultraviolet spectrophotometry on manganese peroxidase (MnP), laccase (Lac), carboxymethyl cellulose (CMCase) and filter paper (FPA). The whole genome was sequenced by Illumina MiSeq and 454 GS Junior platforms. The protein sequences were annotated from the whole genome and compared with COG and KEGG databases through BLASTp to determine several potential lignocellulose-degrading enzymes and pathways. Some of the annotated genes were further verified by realtime RT-PCR. [Results] We obtained strain S12 which was identified as Raoultella ornithinolytica. The bacterium grew to stationary phase after being incubated in CMC-Na liquid medium for 28 h, at which its cellulose degradation related enzymatic activities reached to peak values. Bioinformatic analysis results showed that strain S12 has some significant genes that encode enzymes working in the lignin degradation pathway, such as peroxidase, Fe-Mn superoxide dismutase, catechol 1,2-dioxygenase, protocatechuate 3, 4-dioxygenase, etc. The expression levels of these genes were higher when strain S12 was grown in a medium with lignin as the unique carbon source than in a medium with glucose as the unique carbon source. Also, strain S12 has a complete cellulose degradation and ethanol generation pathway. [Conclusion] Raoultella ornithinolytica S12 has the ability to degrade lignocellulose effectively, which is significant in promoting the development of the lignocellulose application industry.
Key words: lignocellulose degrading bacteriumscreeningRaoultellawhole-genomemetabolic pathway
木质纤维素是木质素、纤维素以及半纤维素组成的生物质,是木本、草本植物难以被降解的细胞壁主要组成成分,其中纤维素作为骨架,木质素与半纤维素以包容物的形式分散在纤维素之中及其周围,三者结合十分稳定且不易降解,它是目前生物质能源研究和利用的主要材料。木质素是造纸工业的副产物,木质素成分几乎完全(90%-95%)溶解在黑液中,这些废水若不经过处理而被直接排放到环境中,由于很难被降解的深色可溶性木质素化合物的存在,会导致严重的水生态系统污染问题[1],而采用化学试剂处理这些废弃物会造成二次污染,因此,有必要开发低能耗、高效率和对环境友好的生物法降解木质纤维素。当前,有很多文献报道了多种能降解木质纤维素的细菌,国内外在这方面的研究也越来越多,来自印度的一个研究团队从造纸废水中分离出10余株具有木质纤维素降解能力的细菌,在应用于造纸黑液处理时都展现出了良好的效果[2, 3]。研究者们从造纸废水或森林土壤中筛选鉴定出不同种属的细菌,如Aneurinibacillus aneurinilyticus[4]、Bacillus sp.[5]、Klebsiella sp.BRL6-2[6]、Sphingomonas paucimobilis SYK-6[7]、Rhodococcus jostii RHA1[8],初步证实它们具有打开木质素联苯结构的功能。越来越多的研究也表明,细菌在某些方面比真菌更具优势,如环境适应性和生物多样性,利用细菌降解木质纤维素,开发出商业化的生物技术具有更广阔的应用前景[9, 10]。尽管针对木质纤维素的降解方法已经有了几十年的研究,仍然有许多问题还未阐明,如细菌的木质纤维素降解酶学、生物信息学等方面的研究还很少。在此基础上,本研究利用选择性培养基初步筛选具备木质纤维素降解功能的细菌,结合二代测序和基因组完成图分析,进一步明确菌株S12具有以羧甲基纤维素钠或碱性木质素为唯一碳源进行生长代谢的木质纤维素高效潜在降解性能,并对其相关酶学和分子生物学展开研究,为开发出具有实际应用价值的木质纤维素生物降解技术奠定基础。
1 材料和方法 1.1 菌种来源 在南京市紫金山选择部分腐木以及枝叶覆盖区为采集点,采集腐木块和枝叶覆盖区的土壤。
1.2 培养基 不同碳源培养基见表 1。其中,培养基1和2中的组分一与组分二共同灭菌,培养基3、4和5中的组分一和所有培养基中的组分三需单独经过0.22 μm水系滤膜过滤灭菌。配制过程中使用无菌水定容,自然pH。
表 1. 不同碳源培养基Table 1. Culture media with different carbon sources
| Media | 1 | 2 | 3 | 4 | 5 |
| Component 1 | CMC-Na 10 g/L | Lignin alkali 1.0 g/L | Azure-B 25 mg/L | Phenol red 10 g/L | Guaiacol 9 mmol/L |
| Component 2 | BM* | ||||
| Component 3 | Cycloheximide 25 mg/L | ||||
| *Basic medium: (NH4)2SO4 2 g/L, K2HPO4 1 g/L, KH2PO4 1 g/L, MgSO4 0.2 g/L, MnSO4 0.02 g/L, Yeast extract 1 g/L (Peptone 10 g/L). | |||||
表选项
1.3 样品处理和初筛 将收集的腐木块剪碎,同时细化土壤,分别称取10 g放入装有90 mL 0.7% NaCl的三角瓶中,盖上封口膜,摇瓶后静置,转移上清并继续静置。取上清100 μL,加入900 μL 0.7% NaCl中,梯度稀释到10-7。每个梯度各取100 μL涂布于培养基3上,同时设置3个平行,封口膜封口,30 ℃避光培养(下同)。
复筛:从培养基3上所有菌落初步挑选N个分别接种于培养基4和5上培养,观察形态。培养基3、4中的Azure-B[11]、Phenol red[3]可作为染料检测过氧化物酶的产生与否,培养基5中的Guaiacol[12]可检测漆酶的表达情况。根据菌落周围的脱色圈大小及菌落颜色变化程度可以比较它们生长能力差异,进一步筛选出优势菌株M株。同时将所筛菌株接种至培养基1培养5 d,用1 mg/mL刚果红(Congo red,CR)染色菌落10 min左右,观察脱色圈大小,以判断纤维素相关酶活高低,比较差异后得到优势菌株K株。
1.4 菌株纯化用平板划线法将所筛菌株分别接种到培养基2上,纯化10次获得纯菌株后于Luria Broth(LB)中培养至对数期,用甘油保种,存放在-80 ℃。
1.5 菌种鉴别菌株经过LB培养至对数期后,使用Bacterial DNA Kit(Omega)提取gDNA,以gDNA为模板,使用27F/1492R通用引物进行16S rRNA基因PCR。为排除gDNA不纯对测序结果的影响,本研究使用pMD19-T Vector Cloning Kit(TaKaRa)以及E.coli DH5α感受态细胞对细菌16S rRNA基因序列进行克隆实验。挑取含0.1%氨苄青霉素平板上的菌落45个,使用M13-47/RV-M通用引物对菌体进行16S rRNA基因PCR,根据电泳条带有无,挑选其中阳性克隆菌落,培养增殖后使用Plasmid Mini Kit(Omega)分别提取质粒,送上海英骏测16S rRNA基因序列,将序列上传到NCBI数据库中进行BLASTn比对。以所筛菌株16S rRNA基因序列及比对结果中相似度较高的4条16S rRNA基因序列为材料,使用MUSCLE将它们连结起来,结合GTR+G+I模型的MrBayes软件绘制系统发育树,判断菌株S12所属类别。
1.6 木质素降解酶活力测定(1)漆酶(Lac)活力测定根据酶氧化ABTS(2,2-连氮-二-3-乙基苯并噻锉-6-磺酸)速率的方法测定Lac酶活。反应体系是,100 μL 0.1 mol/L乙酸钠缓冲液(pH 5.0);800 μL 0.03% ABTS;用牙签挑取单菌落接种至液体培养基2中培养细菌、离心后取上清100 μL。30 ℃测定420 nm下1 min的吸光值变化。定义每分钟氧化1 μmol的ABTS为一个酶活单位U,消光系数ε=3.6×104/(mol·cm)。(2)锰过氧化物酶(MnP)活力测定根据酶在H2O2存在下将Mn2+氧化成Mn3+的速度的方法测定MnP酶活。反应体系是,600 μL 50 mmol/L乳酸钠缓冲液(pH 4.5);100 μL 0.01%酚红;50 μL 2 mmol/L MnSO4;200 μL 0.5% BSA;液体培养基2培养细菌、离心后取上清100 μL。加入50 μL 2 mmol/L H2O2启动反应,30 ℃测定610 nm下1 min的吸光值变化。定义每分钟氧化1 μmol的Mn2+成Mn3+为一个酶活单位U,消光系数ε=2.2×104/(mol·cm)[13, 14]。空白样品测定过程中使用培养基2代替上清液,对应体系中其余成分相同。酶活计算公式[15]:
 ,单位U/L,其中,V总和V(酶)分别代表酶活测定反应体系的总体积及反应添加的上清液体积。
,单位U/L,其中,V总和V(酶)分别代表酶活测定反应体系的总体积及反应添加的上清液体积。1.7 纤维素降解酶活力测定(1)标准曲线绘制及滤纸酶(FPA)活力测定参照GB/T 23881-2009[16],待测样品为用牙签挑取单菌落接种至液体培养基1中生长到不同时期的菌液离心后的上清。(2)羧甲基纤维素酶(CMCase)活力测定称取2.0 g CMC-Na溶于200 mL蒸馏水中,加入0.1 mol/L(pH 4.6)醋酸钠缓冲液100 mL,混匀即为CMC-Na溶液。取1.5 mL CMC-Na溶液与0.5 mL待测上清液于40 ℃水浴保温30 min后立即加1.5 mL DNS显色剂,沸水浴5 min,取出立即冷却,在540 nm下测OD值。以1 mL液体酶在一定条件下,每分钟水解CMC-Na产生1.0 μg的葡萄糖为1个酶活单位,用U/g(U/mL)表示。空白样以0.5 mL培养基1代替酶液。根据OD值从标准曲线上查得葡萄糖含量P。酶活计算公式:P×103/V×T,其中,V代表反应中上清液体积,T代表反应时间。
1.8 全基因组测序和分析细菌完整基因组测序与拼接参见作者所发表文章[17]。使用BLASTp[18]工具,借用目前运用最广泛的代谢通路数据库KEGG(Kyoto Encyclopedia of Genes and Genomes)和蛋白质功能数据库COG(Clusters of Orthologous Groups)与该基因组预测的氨基酸序列逐一进行比对(要求单个序列与COG某一序列匹配数量>90%,匹配质量>20%),分别对其生物学代谢通路和蛋白质功能进行分类。另外,选取NCBI细菌全基因组数据库中全部拉乌尔菌属以及大部分克雷伯氏菌属的细菌全基因组及基因集,下载并与菌株S12的全基因组序列及基因集比较,绘制系统进化树,判断菌株S12所属类别。
1.9 定量RT-PCR验证收集分别在以1 g/L碱性木质素和1 g/L葡萄糖为唯一碳源的培养基中生长至24 h的S12菌体,用RNeasy Protect Bacteria Mini Kit(Qiagen)提取总RNA,再用M-MLV Reverse Transcriptase(Invitrogen)进行反转录合成cDNA备用。使用NCBI网站的Primer BLAST工具,基于S12基因组序列设计目的基因和内参16S rRNA基因的RT-PCR引物序列,由上海捷瑞生物工程有限公司合成,其序列及产物长度见表 2。含目的基因的质粒标准品的制备参照上述“菌种鉴别”相关步骤。本研究使用QuantiNova SYBR Green PCR Kit(Qiagen)配合Applied Biosystems StepOne Plus Real-Time PCR System,进行两步法RT-PCR扩增,所获得的到达阈值时的循环数Ct值由StepOne Plus software 2.2.2收集并用ΔΔCt法分析计算不同样品相应基因的表达量。
表 2. 定量RT-PCR所需引物Table 2. Primers in realtime RT-PCR
| ORF | Forward primer sequence (5′→3′) | Reverse primer sequence (5′→3′) | Product length/bp |
| 16S rRNA gene | GGAGGAAGGCGATAAGGTT | ACATCCGACTTGACAGACC | 160 |
| TE10_02730 | TCGGCTCCGTTGATAACTTC | GATAGCTTCACCCATCAGCG | 149 |
| TE10_13335 | TGAATACGCGGATGTACTTTG | CCTGCAGATGAATATCGAACC | 156 |
| TE10_13810 | AGCCGTGCAGAAATTATTACG | CTCGTGGAGATAGTTCACCG | 166 |
| TE10_13815 | GTGCCTCTATCCTAACGTCT | GAAATCCTCGTACTGGCGAA | 163 |
| TE10_14035 | CGACCCGGAATTCGATAAGA | ATCAGGTCTTCTTTCGGGAC | 153 |
| TE10_15215 | GACCTGTCACATCAACTACG | AGTTCCACTTGATCCGTCC | 137 |
| TE10_23180 | GACGGTGATCGTTAGCCAG | ATCTCAAAGGATTTCCCGGC | 144 |
表选项
2 结果和分析 2.1 木质纤维素降解菌的复筛结果 在木质纤维素降解菌的分离筛选实验中,根据菌落在含有不同木质素类似物培养基上脱色/染色圈的大小定性判断菌株利用碳源的能力。使用CMC-Na培养基以及刚果红染色的方法检测菌株降解纤维素能力。从Azure-B平板上所有菌落中初步筛选50(N)个分别接种于培养基4和5上并观察形态,进一步筛选出优势菌株40(M)株。同时将所筛菌株接种至CMC-Na平板上,经CR染色后比较脱色圈大小,最终选取11(K)株,分别命名各菌株为ZJ18,ZJ15,ZJ8,ZJ5,ZJ44,S12,ZJ17,ZJ19,ZJ32,ZJ7,ZJ21。在不同培养基上点种这些菌株后观察生长到5 d时菌落的大小/脱色圈直径,结果见表 3,其中,菌株S12和菌株ZJ18分别在CMC-Na平板和Azure-B平板上的脱色情况见图 1。根据实验结果可以判断,菌株S12在培养基2和3上的生长速度皆快于其他菌株,选取S12进行全基因组测序和分析。
表 3. 菌落大小/脱色圈直径及菌种分类Table 3. Sizes of colonies/decoloration circle diameters and bacterial classifications
| Strains | Decoloration circle diameter on medium 1 with CR/cm | Size of bacterialcolonies on different mediums | Bacterial classifications according to 16S rRNA gene | |||
| 2 | 3 | 4 | 5 | |||
| ZJ15 | 4.5+0.2 | ++ | + | + | +++ | Bacillus |
| ZJ 5 | 3.8+0.2 | ++ | ++ | + | – | Lysobacter |
| ZJ 8 | 3.2+0.2 | +++ | + | ++ | +++ | Streptomyces |
| ZJ 17 | 3.1+0.2 | ++ | ++ | + | + | Xanthomonas |
| S12 | 1.8+0.2 | +++ | +++ | ++ | ++ | Raoultella |
| ZJ 18 | 1.7+0.2 | +++ | +++ | ++ | ++ | Raoultella |
| ZJ 32 | 1.7+0.2 | ++ | ++ | + | + | Ensifer |
| ZJ 7 | 1.6+0.2 | ++ | ++- | + | + | Mitsuaria |
| ZJ 19 | 1.2+0.2 | ++ | +++ | +++ | +++ | Acinetobacter |
| ZJ44 | 1.1+0.2 | ++ | ++ | + | ++ | Stenotrophomonas |
| ZJ 21 | 1.0+0.2 | + | ++ | + | + | Pseudomonas |
| +: positive ; –: negative. | ||||||
表选项
 |
| 图 1. 菌株S12和菌株ZJ18脱色情况 Figure 1. Decoloration of strains S12 and ZJ18 |
| 图选项 |
2.2 菌株S12的菌种鉴定 将11株菌的16S rRNA基因序列与2014年9月的NCBI中现有数据库进行BLASTn序列比对分析,选取结果中相似度最高的菌属作为对应菌株分类标准,绘制系统进化树。由图 2的A和B可知,菌株S12的16S rRNA基因序列与登录号为KJ599629的拉乌尔菌属菌株16S rRNA基因序列相似度为99%,同时,其全基因组序列与登录号为NC021066的拉乌尔菌属菌株亲缘关系最为相近,因此鉴定菌株S12为解鸟氨酸拉乌尔菌(Raoultella ornithinolytica)。
 |
| 图 2. 依据16S rRNA基因序列(A)和全基因组核心基因(B)构建的菌株S12的系统进化树 Figure 2. Phylogenetic tree of strain S12 based on 16S rRNA gene sequences (A) and core gene sequences (B). The GenBank accession numbers are given in parentheses. The phylogenetic tree was constructed by MrBayes using random concatenation of aligned core genes as the dataset and GTR+G+I as the substitution model. The chain length was set to 10000000 (1 sample/1000 generations) whilst the burn-in was set as 2000. Posterior probabilities are denoted at nodes. Numbers at branching points are bootstrap values. Bar 0.02 (0.05) at the bottom is the sequence divergence. |
| 图选项 |
2.3 菌株S12相关酶活比较2.3.1细菌生长曲线:S12在液体培养基1和2中的菌体(包括活菌和死菌)密度用OD600表示,其变化趋势如图 3所示,分别培养至28 h和1 d时,菌体生长达到稳定期。
 |
| 图 3. 菌株S12生长曲线 Figure 3. Growth curve of strain S12. A: Carbon source: CMC-Na; B: Carbon source: lignin alkali |
| 图选项 |
2.3.2纤维素降解酶活力的变化: 一般认为,滤纸酶活主要反映了由外切酶(EC:3.2.1.9)、内切酶(EC:3..2.1.4)和β-糖苷酶(EC:3.2.1.21)3种纤维素酶所组成的诱导复合酶系协同/竞争降解纤维素的能力[19]。在以CMC-Na为碳源的培养过程中,菌株S12的CMCase和FPA的酶活力变化如图 4-A所示。从图中看出培养初始阶段酶活力较低,在28 h左右时,FPA酶活力达到最大值1.43 U/mL,而在32 h左右时,CMCase酶活力达到最大值1.03 U/mL,相对Irfan等[20]筛选的Cellulomonas sp.ASN2的CMCase最高酶活力(0.43 U/mL)较高。推测,在细菌生长过程中,诱导复合酶系的酶活力大小与活菌密度呈正比,其酶活力高峰值比CMCase提前一段时间,这可能由菌群利用碳源的策略来决定。
 |
| 图 4. 菌株S12的木质纤维素降解酶活性 Figure 4. Lignocellulose degradation enzyme activity of strain S12. A: Carbon Source: CMC-Na; B: Carbon source: lignin alkali. |
| 图选项 |
2.3.3木质素降解酶活力的变化:在培养基2的生长过程中,菌株S12的MnP和Lac的酶活力变化如图 4-B所示,从图中看到培养初期酶活力较低,在对应生长稳定期的第2.5天,菌株S12的MnP和Lac的酶活力分别为31.58 U/L和23.21 U/L,前者与Kapich等[21]测定的Phanerochaete chrysosporium ME-446最高酶活力(41.4 U/L)相当。酶活曲线的峰点出现时间比Chen等[22]测定的Novosphingobium sp.B-7两种酶活力提前3 d,但比菌株S12生长曲线的峰点出现时间晚1.5 d,这可能是由于细菌在生长的次生代谢阶段合成木质素降解酶类的速度加快,在有限的生长条件下,细菌生长达到停滞或者衰亡期时,其合成并释放到细胞外的酶类减少,细菌降解木质素能力减弱。MnP在Mn2+环境中,由H2O2催化氧化酚型结构的木质素,如木质素结构单体香豆醇、松柏醇和芥子醇。漆酶利用分子氧作为氧化物,可将酚环氧化成苯氧自由基,并攻击木质素的苯环结构,从而降解木质素。有研究报道在多种细菌中检测到了漆酶的活性[23, 24, 25],虽然从细菌中克隆、测序和表达漆酶编码基因的报道较少,但仍然得到了一些确切的结果[26, 27],证实细菌也可以分泌漆酶并具有相应的编码基因。在本研究中,从菌株S12的培养液中检测到了漆酶活性,这表明菌株S12中存在漆酶或多酚氧化酶编码基因。将菌株S12基因组CDS序列对应的氨基酸序列与相关漆酶基因氨基酸序列进行BLASTp比对后,在匹配长度大于90%的基础上,发现基因组中标签为TE10_06395的序列与芽孢杆菌属的2个cotA基因(GenBank登录号分别为JQ734990和JQ797387)相似度大于25%,与链霉菌属的Lac(EC:1.10.3.2)的氨基酸序列相似度大于23%。
2.4 全基因组测序结果 菌株S12在GenBank上的登录号为CP010557。该菌基因组全长为5522044 bp,(G+C)%含量为57.47%,不包含质粒。
2.5 基因注释和分类2.5.1COG功能分类:根据PGAAP对基因组的注释结果,得到5075个已知基因序列,预测到4875个CDS(coding sequences)。在与COG数据库BLASTp比对之后,有78.4%的CDS序列得到功能分类,如图 5所示,其中具备次生代谢产物生物合成、运输和分解代谢(Q)功能的基因共有128个,具备细胞内物质转运、分泌和膜泡运输(U)功能的基因有84个,具备碳水化合物的运输和代谢(G)功能的基因共有553个,这些基因可能与该细菌降解碱性木质素和羧甲基纤维素钠的功能密切相关。
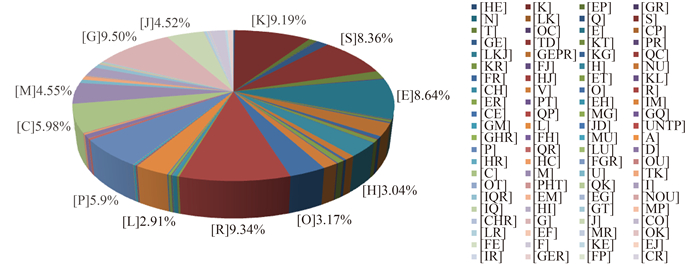 |
| 图 5. 菌株S12基因组功能分类 Figure 5. COG class annotation distribution of strain S12 genome. The COG-annotated genes are grouped under their respective COG classes. Only their class abbreviations are used in this graph, their corresponding class descriptions are listed as below: N, cell motility; Q, secondary metabolite biosynthesis, transport and catabolism; U, intracellular trafficking, secretion, and vesicular transport; D, cell cycle control, cell division, chromosome partitioning; T, signal transduction mechanisms; I, lipid transport and metabolism; H, coenzyme transport and metabolism; V, defense mechanisms; O, posttranslational modification, protein turnover, chaperones; C, energy production and conversion; F, nucleotide transport and metabolism; P, inorganic ion transport and metabolism; M, cell wall/membrane/envelope biogenesis; K, transcription; L, replication, recombination and repair; E, amino acid transport and metabolism; G, carbohydrate transport and metabolism; S, function unknown; R, general function prediction only; J, translation, ribosomal structure and biogenesis. Percentages of the top twelve classes are labeled for easy reference. |
| 图选项 |
2.5.2KEGG代谢通路分类:根据KEGG对该菌株的分析结果得知,在近196种代谢通路中,淀粉和蔗糖代谢通路包括了33个基因,参与芳香化合物代谢的基因有29个。其中,有2种过氧化物酶(EC:1.11.1.21)(EC:1.11.1.9)分别存在于苯丙素生物合成和花生四烯酸等化合物的代谢过程中,产H2O2酶辅助参与木质素的降解,即为木质素过氧化物酶和锰过氧化物酶提供H2O2。香草醛是木质素降解过程中重要的中间产物之一,香草醛脱甲基酶(EC:1.14.13.82)能将香草醛脱甲基化变成原儿茶酸[28]。邻苯二酚-1,2-双加氧酶(EC:1.13.11.1)和原儿茶酸-3,4-双加氧酶(EC:1.13.11.3)能使邻苯二酚和原儿茶酸这2种木质素降解的中间产物开环裂解成脂肪族物质进入三羧酸循环[29],它们的CDS序列号见表 4。经过定量RT-PCR验证,所查找到的与木质素单体降解相关的基因,在以碱性木质素为碳源的培养条件下的表达量不同程度地高于以葡萄糖为碳源的培养条件,说明碱性木质素环境能够诱导这些基因达到较高的表达水平,以保证菌株S12有效利用环境中的碳源,其结果见图 6。由于部分重要的木质素降解酶如漆酶,大多数都是在霉菌中被发现,细菌中的报道相对较少,推测数据库中暂未记录此类细菌漆酶的相关数据,因此在具有碱性木质素降解功能的菌株S12的代谢通路分析中未出现漆酶。同时,鸟氨酸氨甲酰基转移酶(EC:2.1.3.3)、纤维素合成酶(EC:2.4.1.12)和羧甲烯丁烯羟酸内酯酶(EC:3.1.1.45)也出现在菌株S12的基本代谢通路注释结果中。这些基因和通路的存在,为菌株S12降解木质纤维素提供了依据。深入分析KEGG所注释的菌株S12代谢通路,我们得到淀粉和蔗糖及糖酵解等通路中参与生成乙醇的酶的相关信息,从而绘制出纤维素的降解和乙醇生成途径,如图 7所示,其中所参与的酶及对应CDS序列号见表 5。
表 4. 菌株S12基因组中木质素降解酶类Table 4. Lignin-degrading enzymes in strain S12 genome
| Lignin-degrading enzymes | Definitions | Locus_tags (CDS) | Orthologous genes |
| Peroxidases | Catalase-peroxidase [EC:1.11.1.21] | TE10_14035 | K03782 |
| Glutathione peroxidase [EC:1.11.1.9] | TE10_15215 | K00432 | |
| Hydrogen peroxide-producing enzymes | Superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | TE10_02730 | K04564 |
| Catechol 1,2-dioxygenase [EC:1.13.11.1] | TE10_13810 | K03381 | |
| Biphenyl ring cleavage enzymes | Protocatechuate 3, 4-dioxygenase, alpha subunit [EC:1.13.11.3] | TE10_13335 | K00448 |
| Benzoate/toluate 1,2-dioxygenase alpha subunit [EC:1.14.12.10 1.14.12.-] | TE10_13815 | K05549 | |
| Vanillate O-demethylase ferredoxin subunit [EC:1.14.13.82] | TE10_23180 | K03863 |
表选项
表 5. 菌株S12的纤维素降解和乙醇生成途径中的酶Table 5. Enzymes in cellulose-degrading and ethanol-producing pathway of strain S12
| Enzymes | Definitions | Locus_tags (CDS) | Orthologous genes |
| 1 | Starch phosphorylase [EC:2.4.1.1] | TE10_00530 | K00688 |
| 2 | PTS system, fructose-specific IIA component [EC:2.7.1.69] | TE10_11530 | K02768 |
| 3 | Beta-fructofuranosidase [EC:3.2.1.26] | TE10_08465 | K01193 |
| 4 | Fructokinase [EC:2.7.1.4] | TE10_01710 | K00847 |
| 5 | Glucose-6-phosphate isomerase [EC:5.3.1.9] | TE10_03680 | K01810 |
| 6 | Phosphoglucomutase [EC:5.4.2.2] | TE10_09370 | K01835 |
| 7 | UTP--glucose-1-phosphate uridylyltransferase [EC:2.7.7.9] | TE10_17490 | K00963 |
| 8 | Cellulose synthase (UDP-forming) [EC:2.4.1.12] | TE10_01065 | K00694 |
| 9 | Endoglucanase [EC:3.2.1.4] | TE10_01055 | K01179 |
| 10 | Beta-glucosidase [EC:3.2.1.21] | TE10_12500 | K05349 |
| 11 | Glucokinase [EC:2.7.1.2] | TE10_02625 | K00845 |
| 12 | Glucose-6-phosphate isomerase [EC:5.3.1.9] | TE10_03680 | K01810 |
| 13 | 6-phosphofructokinase 1 [EC:2.7.1.11] | TE10_02760 | K00850 |
| 14 | Fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | TE10_07505 | K01624 |
| 15 | Triosephosphate isomerase (TIM) [EC:5.3.1.1] | TE10_01690 | K01803 |
| 16 | Glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] | TE10_12115 | K00134 |
| 17 | Phosphoglycerate kinase [EC:2.7.2.3] | TE10_23680 | K00927 |
| 18 | 2, 3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | TE10_01480 | K15633 |
| 19 | Enolase [EC:4.2.1.11] | TE10_22565 | K01689 |
| 20 | Pyruvate kinase [EC:2.7.1.40] | TE10_17205 | K00873 |
| 21 | Pyruvate dehydrogenase E1 component alpha subunit [EC:1.2.4.1] | TE10_14430 | K00161 |
| 22 | Pyruvate dehydrogenase E1 component beta subunit [EC:1.2.4.1] | TE10_14425 | K00162 |
| 23 | Pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12] | TE10_06345 | K00627 |
| 24 | Dihydrolipoamide dehydrogenase [EC:1.8.1.4] | TE10_06350 | K00382 |
| 25 | Phosphate acetyltransferase [EC:2.3.1.8] | TE10_20305 | K13788 |
| 26 | Acetate kinase [EC:2.7.2.1] | TE10_20300 | K00925 |
| 27 | Aldehyde dehydrogenase (NAD+) [EC:1.2.1.3] | TE10_20355 | K00128 |
| 28 | Alcohol dehydrogenase [EC:1.1.1.1] | TE10_02675 | K00001 |
表选项
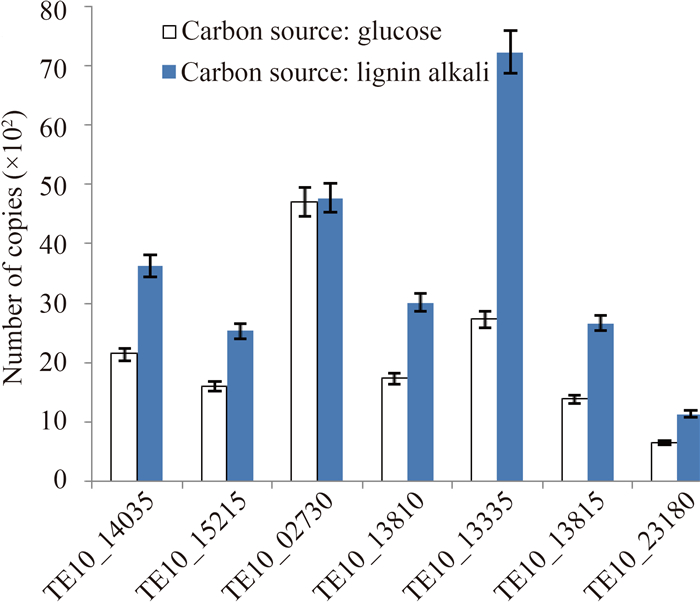 |
| 图 6. 菌株S12相关基因在两种碳源下差异表达量 Figure 6. Relative genes expression of strain S12 under two different carbon sources. |
| 图选项 |
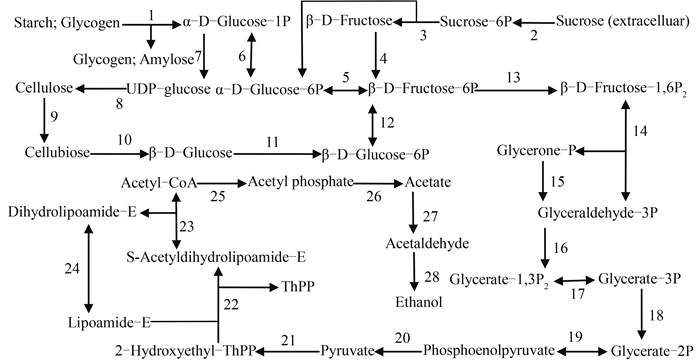 |
| 图 7. 菌株S12的纤维素降解和乙醇生成途径 Figure 7. Cellulose-degrading and ethanol-producing pathway of strain S12. |
| 图选项 |
3 讨论 木质素过氧化物酶(LiP)、锰过氧化物酶(MnP)以及漆酶(Lac)这3种酶在木质素降解过程中的作用主要是攻击木质素聚合物结构,将其解聚成更小的分子或者单体的苯环物质。Raoultella ornithinolytica S12基因组中TE10_06395的序列与芽孢杆菌属的2个cotA基因相似度大于25%,与链霉菌属的Lac氨基酸序列相似度大于23%,该段序列所编码的蛋白在菌株生长过程中表现的功能可能与漆酶相当。部分研究表明,细菌在木质素单体苯化合物降解方面表现出较强的能力,过氧化物酶、芳环开裂酶等酶系在木质纤维素的降解过程中所发挥的作用和功能已被证实[30, 31, 32, 33, 34, 35]。Raoultella ornithinolytica S12分别在以愈创木酚和酚红为唯一碳源的培养基中生长良好,证明该菌能够较好地利用酚型木质素单体物质。生物信息学分析结果表明,该菌具有木质素降解通路中重要酶类的编码基因,如过氧化物酶、Fe-Mn型超氧化物歧化酶、邻苯二酚1,2-双加氧酶、原儿茶酸-3,4-双加氧酶和香草酸脱甲基酶,这些基因在以碱性木质素为碳源的条件下培养24 h的表达量不同程度地高于以葡萄糖为碳源的培养条件。
由近年的宏基因组学研究可知,森林土壤和腐烂木材中具有丰富的微生物多样性,其中很多类型的微生物可能降解植物主要部分并将营养成分释放到环境中,从而参与养分循环。如果能够有效开发微生物降解木质纤维素的催化通路,木质纤维素将会成为一种丰富的可再生芳香化学品以及生物燃料乙醇的潜在来源。全基因组测序及分析为揭示新型木质纤维素降解菌代谢途径和调控机制提供可能,本研究首次揭示了Raoultella ornithinolytica S12具有效的木质纤维素降解性能,其纤维素降解和乙醇生成通路已经初步明确,同时存在几种重要的木质素降解酶编码基因,从基因层面证实了该菌具有木质纤维素降解能力,为构建生产木质纤维素降解酶的基因工程菌提供了有价值的依据。利用Raoultella ornithinolytica S12有望能开发出可商业化应用的木质纤维素生物降解技术,通过降解木质纤维素生成有价值的化工产品,或者用于处理造纸黑液等木质素造成的环境污染问题,还可应用于生产如香草酸等有价值的芳香化学品。
致谢
感谢南京农业大学生物信息学中心许志晖对本研究的支持和帮助。
参考文献
| [1] | Berryman D, Houde F, DeBlois C, O'hea M. Nonylphenolic compounds in drinking and surface waters downstream of treated textile and pulp and paper effluents:a survey and preliminary assessment of their potential effects on public health and aquatic life. Chemosphere, 2004, 56(3):247-255. |
| [2] | Chandra R, Abhishek A, Sankhwar M. Bacterial decolorization and detoxification of black liquor from rayon grade pulp manufacturing paper industry and detection of their metabolic products. Bioresource Technology, 2011, 102(11):6429-6436. |
| [3] | Chandra R, Raj A, Purohit HJ, Kapley A. Characterisation and optimisation of three potential aerobic bacterial strains for kraft lignin degradation from pulp paper waste. Chemosphere, 2007, 67(4):839-846. |
| [4] | Raj A, Chandra R, Reddy MMK, Purohit HJ, Kapley A. Biodegradation of kraft lignin by a newly isolated bacterial strain, Aneurinibacillus aneurinilyticus from the sludge of a pulp paper mill. World Journal of Microbiology and Biotechnology, 2007, 23(6):793-799. |
| [5] | Raj A, Reddy MMK, Chandra R, Purohit HJ, Kapley A. Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation, 2007, 18(6):783-792. |
| [6] | Woo HL, Ballor NR, Hazen TC, Fortney JL, Simmons B, Davenport KW, Goodwin L, Ivanova N, Kyrpides NC, Mavromatis K, Woyke T, Jansson J, Kimbrel J, DeAngelis KM. Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2. Standards in Genomic Sciences, 2014, 9:19. |
| [7] | Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Bioscience, Biotechnology, and Biochemistry, 2007, 71(1):1-15. |
| [8] | Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TDH. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry, 2011, 50(23):5096-5107. |
| [9] | Raj A, Reddy MMK, Chandra R. Identification of low molecular weight aromatic compounds by gas chromatography-mass spectrometry (GC-MS) from kraft lignin degradation by three Bacillus sp.. International Biodeterioration & Biodegradation, 2007, 59(4):292-296. |
| [10] | Raj A, Reddy MMK, Chandra R. Decolourisation and treatment of pulp and paper mill effluent by lignin-degrading Bacillus sp. Journal of Chemical Technology and Biotechnology, 2007, 82(4):399-406. |
| [11] | Bandounas L, Wierckx NJP, de Winde JH, Ruijssenaars HJ. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnology, 2011, 11:94. |
| [12] | Wang YX, Liu Q, Yan L, Gao YM, Wang YJ, Wang WD. A novel lignin degradation bacterial consortium for efficient pulping. Bioresource Technology, 2013, 139:113-119. |
| [13] | Chen YH, Chai LY, Zhu YH, Yang ZH, Zheng Y, Zhang H. Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. Journal of Applied Microbiology, 2012, 112(5):900-906. |
| [14] | Bonugli-Santos RC, Durrant LR, da Silva M, Sette LD. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzyme and Microbial Technology, 2010, 46(1):32-37. |
| [15] | Lin JF, Liu ZM, Chen XY, Guo LQ, Wang J. Evaluation of assay methods for determining fungal laccase activity. Chinese Journal of Bioprocess Engineering, 2009, 7(4):1-8. (in Chinese)林俊芳, 刘志明, 陈晓阳, 郭丽琼, 王杰. 真菌漆酶的酶活测定方法评价. 生物加工过程, 2009, 7(4):1-8. |
| [16] | 中华人民共和国国家质量监督检验检疫总局, 中国国家标准化管理委员会. GB/T 23881-2009饲用纤维素酶活性的测定:滤纸法. 北京:中国标准出版社, 2009. |
| [17] | Bao WY, Zhou Y, Jiang JW, Xu ZH, Hou LY, Leung FCC. Complete genome sequence of Raoultella ornithinolytica strain S12, a lignin-degrading bacterium isolated from forest soil. Genome Announcements, 2015, 3(2):e00104-15. |
| [18] | Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Research, 1997, 25(17):3389-3402. |
| [19] | Lynd LR, Wenmer PJ, Van Zyl WH, Pretorius IS. Microbial cellulose utilization:fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 2002, 66(3):506-577. |
| [20] | Irfan M, Safdar A, Syed Q, Nadeem M. Isolation and screening of cellulolytic bacteria from soil and optimization of cellulase production and activity. Turkish Journal of Biochemistry, 2012, 37(3):287-293. |
| [21] | Kapich AN, Prior BA, Botha A, Galkin S, Lundell T, Hatakka A. Effect of lignocellulose-containing substrates on production of ligninolytic peroxidases in submerged cultures of Phanerochaete chrysosporium ME-446. Enzyme and Microbial Technology, 2004, 34(2):187-195. |
| [22] | Chen YH, Chai LY, Tang CJ, Yang ZH, Zheng Y, Shi Y, Zhang H. Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process. Bioresource Technology, 2012, 123:682-685. |
| [23] | Claus H. Laccases and their occurrence in prokaryotes. Archives of Microbiology, 2003, 179(3):145-150. |
| [24] | Sharma P, Goel R, Capalash N. Bacterial laccases. World Journal of Microbiology and Biotechnology, 2007, 23(6):823-832. |
| [25] | Alexandre G, Zhulin IB. Laccases are widespread in bacteria. Trends in Biotechnology, 2000, 18(2):41-42. |
| [26] | Ridge JP, Lin M, Larsen EI, Fegan M, McEwan AG, Sly LI. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environmental Microbiology, 2007, 9(4):944-953. |
| [27] | Wu J, Kim KS, Lee JH, Lee YC. Cloning, expression in Escherichia coli, and enzymatic properties of laccase from Aeromonas hydrophila WL-11. Journal of Environmental Sciences, 2010, 22(4):635-640. |
| [28] | Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. A tetrahydrofolate-dependent O-demethylase, ligM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. Journal of Bacteriology, 2005, 187(6):2030-2037. |
| [29] | Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, Haraguchi T. The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Letters, 1988, 233(1):129-133. |
| [30] | Leonowicz A, Cho NS, Luterek J, Wilkolazka A, Wojtas-Wasilewska M, Matuszewska A, Hofrichter M, Wesenberg D, Rogalski J. Fungal laccase:properties and activity on lignin. Journal of Basic Microbiology, 2001, 41(3/4):185-227. |
| [31] | Nishikawa S, Katayama Y, Yamasaki K, Morohoshi N, Haraguchi T. Cloning and expression of Pseuidomonas paiucimobilis SYK-6 genes involved in the degradation of vanillate in P. putida. Mokuzai Gakkaishi, 1989, 35(2):158-163. |
| [32] | Maruyama K, Shibayama T, Ichikawa A, Sakou Y, Yamada S, Sugisaki H. Cloning and characterization of the genes encoding enzymes for the protocatechuate Meta-degradation pathway of Pseudomonas ochraceae NGJ1. Bioscience, Biotechnology, and Biochemistry, 2004, 68(7):1434-1441. |
| [33] | Chai L Y, Zhang H, Yang WC, Zhu YH, Yang ZH, Zheng Y, Chen YY. Biodegradation of ferulic acid by a newly isolated strain of Cupriavidus sp. B-8. Journal of Central South University, 2013, 20(7):1964-1970. |
| [34] | Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Current Opinion in Biotechnology, 2011, 22(3):394-400. |
| [35] | Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Natural Product Reports, 2011, 28(12):1871-1872. |
