0 引言
【研究意义】稻瘟病菌(Magnaporthe oryzae)侵染水稻引起的稻瘟病是水稻上的重要病害,每年可造成10%—30%的稻谷产量损失,严重威胁世界各水稻产区粮食生产安全[1]。由于经济的重要性和分子遗传的可操作性,稻瘟病菌-水稻互作机制已成为研究病原菌-寄主植物互作的模式系统[1-3]。对稻瘟病菌致病分子机理的深入了解不仅有助于新的稻瘟病防治策略的研发,有效控制病害流行,而且对其他植物病原真菌的研究及其病害防治具有重要的指导意义。【前人研究进展】稻瘟病菌通过形成一种特异性的侵染结构——附着胞侵入寄主细胞[4]。当分生孢子着落在稻株表面后,在适宜的水分条件下萌发产生芽管,芽管顶端迅速膨大并逐渐分化形成附着胞。成熟的附着胞黑色素化并通过甘油等溶质的积累而产生强大膨压(高达8 MPa)。在膨压的作用下,附着胞产生足够的机械压力驱动稻瘟病菌成功穿透寄主表皮进行侵染,随后鳞茎状的侵染菌丝在细胞内和细胞间扩展。侵染3—5 d后水稻叶片表面形成可见的坏死病斑[1,5-6]。研究表明,cAMP-PKA信号途径调控早期表面信号识别和附着胞分化,外源添加cAMP能诱导分生孢子在亲水表面形成附着 胞[7-12]。Pmk1-MAPK信号途径调控附着胞成熟、穿透及扩展[11-14]。PMK1缺失突变体不能形成附着胞,不能穿透寄主表皮,对寄主致病性完全丧失[13]。Mst12是位于Pmk1下游的转录因子,参与调控侵染钉形成和穿透。Δmst12能形成成熟的附着胞,但不能穿透寄主表皮而完全丧失对寄主的致病性[15-17]。由于MST12不是附着胞发育与成熟所必需,因此研究者们认为还存在其他受Pmk1调控的转录因子参与附着胞的形成。近期,笔者实验室研究发现了一个C2H2型锌指结构转录因子Znf1,其可能位于cAMP-PKA和Pmk1-MAPK信号途径下游,在附着胞介导的稻瘟病菌侵染过程中有重要作用[18]。Δznf1在营养生长、色素沉积以及有性生殖等方面与野生型菌株没有明显的差异,但Δznf1芽管顶端不能分化形成成熟的附着胞,不能穿透寄主表皮,即使是在划伤或针刺的大麦和水稻叶片表面都不能产生病斑。在稻瘟病菌附着胞发育阶段,Znf1受Pmk1调控,并且同Δpmk1能响应外源cAMP刺激类似,外源cAMP或IBMX能诱导Δznf1芽管顶端分化形成囊泡状的膨大结构。但酵母双杂交实验表明Znf1与Pmk1没有直接的互作[18]。【本研究切入点】Znf1下游调控的基因尚不明确,Znf1对附着胞相关基因的调控以及与Pmk1的关系有待挖掘,而且对Znf1作用机制的深入研究能够加深对稻瘟病菌附着胞调控机理的认识。本研究通过高通量测序技术,从转录水平上解析Znf1的调控机制。【拟解决的关键问题】利用RNA-Seq(RNA-sequencing)技术对稻瘟病菌野生菌株Guy11和Δznf1突变体进行表达谱分析和差异表达基因筛选,以期为进一步鉴定和研究受Znf1调控的基因提供基础数据。此外,通过比较Δznf1和Δpmk1中的差异表达基因,筛选受Znf1和Pmk1共同调控的基因,深入了解Znf1与Pmk1的关系以及鉴定新的与稻瘟病菌附着胞形成相关基因,从而为完善稻瘟病菌致病分子机理提供数据。1 材料与方法
1.1 供试材料
RNA-Seq所用的稻瘟病菌野生型Guy11、Δznf1和Δpmk1突变体均为笔者实验室保存或研究获得的菌株。供试菌株在CM液体培养基中振荡培养48 h(28℃,180 r/min)后,2层纱布过滤收集菌丝,经无菌水冲洗数遍后,将水分压干,液氮速冻后储存于-80℃保存备用。每个菌株3个重复。CM培养基配方:10 g·L-1 glucose, 2 g·L-1 peptone,1 g·L-1 yeast extract,1 g·L-1 casamino acids,0.1%(v/v)trace elements,0.1%(v/v)vitamin supplement,6 g·L-1 NaNO3,0.5 g·L-1 KCl,0.5 g·L-1 MgSO4,1.5 g·L-1 KH2PO4,pH 6.5。1.2 RNA提取及质量检测
2015年9月样品组织送至深圳华大基因科技服务中心,RNA-Seq测序在该中心进行。采用Trizol法分别提取各菌株的菌丝样品总RNA,并使用DnaseI去除样品中的DNA杂质。通过1%琼脂糖凝胶电泳,以及NanoDrop ND-2000和Agilent 2100对RNA样品进行浓度、纯度和完整性等检测。OD值标准:OD260/280≥ 1.8,OD260/230≥1.8;RIN(RNA完整系数)≥8,28S/18S≥1。为了提高试验的准确性,将3个生物学重复样品分别提取RNA,然后等量混合作为一个样本用于RNA-Seq。1.3 文库构建及RNA-Seq测序
用带有Oligo(dT)的磁珠富集mRNA,向得到的mRNA中加入适量打断试剂高温条件下使其片断化,再以片断化后的mRNA为模板,合成cDNA,经过磁珠纯化、末端修复、3′末端加碱基A、加测序接头后,进行PCR扩增,从而完成整个文库制备工作。构建好的文库用Agilent 2100 Bioanalyzer和ABI StepOne Plus Real-Time PCR System进行质量和产量检测,文库质控合格后进行Illumina HiSeqTM 2000测序分析,测序反应进行一次。1.4 基因定量分析及差异基因筛选
基因表达量用FPKM(Fragments Per Kilobase of exon per Million fragments mapped)法计算[19]。根据野生菌株Guy11和突变体Δznf1或Δpmk1中基因的FPKM值来比较不同样品间的基因差异表达。在本试验分析中,差异基因筛选标准为错误发现率(false discovery rate,FDR)≤0.001且差异倍数在2倍以上即∣log2(样品间基因表达量的比值)∣≥1。1.5 差异基因GO功能注释及KEGG Pathway富集分析
将所有差异表达基因向Gene Ontology(GO)数据库(http://www.geneontology.org/)的各个term映射,计算每个term的基因数目,然后应用超几何检验,找出与整个稻瘟病菌基因组相比,在差异表达基因中显著富集的GO条目。得到差异基因的GO注释后,利用WEGO软件对差异基因进行GO功能分类统计[20]。Pathway显著性富集分析以KEGG Pathway为单位[21],应用GO富集原理可以得出差异表达基因中显著性富集的Pathway。1.6 实时荧光定量PCR(qRT-PCR)
为了验证RNA-Seq结果的可靠性,随机选择10个差异表达基因进行qRT-PCR检测,引物见表1。通过Trizol法提取CM液体培养基振荡培养48 h的菌丝总RNA,用反转录试剂盒PrimeScript®RT Reagent Kit With gDNA Eraser(TaKaRa)合成cDNA。采用GoTag® qPCR Master Mix(Promega)试剂,一个反应体系为20 μL,3个重复。qRT-PCR反应在BIO-RAD CFX96TM Real-Time System仪上运行。反应程序为:95℃ 30 s,95℃5 s,56℃ 15 s,72℃ 20 s,39个循环。以稻瘟病菌β-tubulin(MGG_00604)为内参基因,通过2-△△CT法计算基因相对表达水平[22]。通过3次独立重复试验计算基因相对表达的平均值和标准差。Table 1
表1
表1本研究qRT-PCR引物
Table 1qRT-PCR primers used in the study
| 名称Name | 序列Sequence (5′-3′) | 名称Name | 序列Sequence (5′-3′) | |
|---|---|---|---|---|
| Tublin-F | CATACGGTGACCTGAACTAC | Tublin-R | CCATGAAGAAGTGCAGACG | |
| 10422-F | TGTCGCCGATCAACACAAGC | 10422-R | CATCCGAGTCTCCAAAGTCC | |
| 01391-F | AGGACCGACACTACCGTTTC | 01391-R | TGCGGTGCAGTTGGCTCAATC | |
| 03825-F | ACCGAAGTCGAGATTGTGCG | 03825-R | CGCGCTTAAAGTAGTTACGG | |
| 09000-F | GGCACGGAGACTACAACTAC | 09000-R | ACTTTCGCCTGGTTGATCAG | |
| 09994-F | CGATCACTCTCGGTATCTTCC | 09994-R | AGGATCGGGAAGAACATGGC | |
| 05719-F | AGGACGCTGAGAAGAACACC | 05719-R | GCTTCTTGGAGACACCCTTC | |
| 03329-F | GCTCTTCCGACTCATTGACG | 03329-R | TCTCCTGCAGGTCGAACTTG | |
| 02246-F | TGAGGAGTCCAAGCAGCAC | 02246-R | TCCTTGGCAGACTCCTTGG | |
| 08944-F | CCTGACCAATGGCAACAAGC | 08944-R | TTGTCGACGGAGCCCTCAA | |
| 11084-F | CCTCCATGGTTCAGGTCAG | 11084-R | GTCTCCTCGTGATTCTTGTC |
新窗口打开
2 结果
2.1 表达谱测序及测序质量评估
为了深入了解稻瘟病菌中受转录因子ZNF1调控的基因,对稻瘟病菌野生型菌株Guy11以及ZNF1基因缺失突变体菌株Δznf1进行表达谱分析。收集菌株Guy11和Δznf1在液体CM培养基中培养2 d的菌丝,并分别进行样品Total RNA抽提。NanoDrop ND-2000及Agilent 2100对Total RNA的质检结果表明,样品RNA的浓度、纯度及完整性等各项指标达到建库测序要求。进而构建测序文库,文库质控合格后利用Illumina HiSeqTM 2000进行测序。测序得到的原始图像数据经base calling转化的序列数据称为raw reads。将低质量序列以及接头序列等杂质raw reads过滤后得到可用于数据分析的clean reads。菌株Guy11和Δznf1分别获得22 100 589和22 074 094个clean reads,分别占原始数据的98.85%和98.72%。对数据质量评估合格后(Q20>95%),将clean reads比对到水稻稻瘟病菌基因组序列(http://www.broadinstitute.org/annotation/genome/magnaporthe_grisea/MultiHome.html)。比对结果显示,Guy11和Δznf1分别有92.63%和92.37%的clean reads能有效的比对到参考基因组上,并且具有唯一比对位点的reads数分别占比对到基因组上总reads数的89.20%和87.88%(表2),表明样品测序质量较高。
Table 2
表2
表2样品RNA-Seq序列与参考基因组的比对统计
Table 2Statistical analysis of RNA-Seq reads mapping to the reference genome
| Reads比对统计 Statistics of reads mapping | Guy11 | Δznf1 | ||
|---|---|---|---|---|
| Reads数目 Reads number | 百分比 Percentage (%) | Reads数目 Reads number | 百分比 Percentage (%) | |
| 总reads Total reads | 22100589 | 100.00 | 22074094 | 100.00 |
| 比对上的reads Total mapped reads | 20471630 | 92.63 | 20389943 | 92.37 |
| 完美匹配的reads Perfect match reads | 17561189 | 79.46 | 17578874 | 79.64 |
| 唯一位点匹配的reads Unique match reads | 19712923 | 89.20 | 19399539 | 87.88 |
| 多位点匹配的reads Multi-position match reads | 758707 | 3.43 | 990404 | 4.49 |
| 未比对上的reads Total unmapped reads | 1628959 | 7.37 | 1684151 | 7.63 |
新窗口打开
测序饱和度在一定程度上可以评估测序数据量是否达到要求。由图1所示,在测序量(reads数量)较小时,样品检测到的基因数均随着测序量的增加而随之上升,当测序量达到5 M左右后,其增长速度趋于平缓,说明检测到的基因数趋于饱和。菌株Guy11和Δznf1均产生不低于20 M的clean reads,因此可以认为测序检测到的基因数能够基本覆盖细胞表达的全部基因,测序量已达到要求。
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1RNA-Seq测序饱和度分析
-->Fig. 1Analysis of sequencing saturation
-->
2.2 Δznf1差异表达基因的筛选
为鉴定受Znf1调控相关基因,对野生型Guy11和Δznf1表达谱进行比较,获得差异表达基因(differentially expressed genes,DEGs)。根据基因的表达量(FPKM值)计算该基因在两个样品间的差异表达倍数,通过控制错误发现率FDR来决定P-value的域值。以∣log2 ratio (Δznf1/Guy11)∣≥1(即倍数差异在2倍以上),并且FDR≤0.001为筛选标准,共获得709个差异表达基因,其中上调表达基因299个,∣log2 ratio∣值>5(即上调倍数大于32)的有22个,下调表达基因410个,∣log2 ratio∣值>5的有12个(图2-A、2-B;附表1)。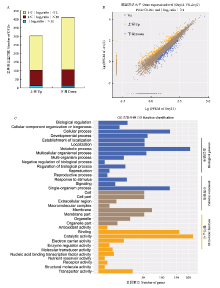 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图2Δznf1差异表达基因分析A:Δznf1差异基因数目统计Statistical analysis of DGEs in Δznf1;B:基因表达量的散点图。横、纵轴分别为Guy11和Δznf1基因表达量的对数值,橙色点表示上调基因,蓝色点表示下调基因,褐色是非显著性差异基因Scatter plot of gene expression. The horizontal and vertical axis were the lg (FPKM) value for all genes in Guy11 and Δznf1, respectively. Orange points represented up-regulated genes, blue points represented down-regulated genes, and brown points represented not significant genes;C:差异基因GO功能分类 GO categories of the DEGs
-->Fig. 2Analysis of differentially expressed genes in Δznf1
-->
Table S1
附表1
附表1Δznf1菌丝中差异表达基因(|log2 ratio|≥1且 FDR<0.001)
Table S1Differentially expressed genes in the mycelia of Δznf1(|log2 ratio|≥1 and FDR<0.001)
| 基因编号 Gene ID | Log2 ratio (znf1/Guy11) | 上调-下调 Up-Down-Regulation | 基因描述 Gene description |
|---|---|---|---|
| MGG_11300 | 16.40789841 | Up | hypothetical protein (213 nt) |
| MGG_15270 | 12.58871464 | Up | predicted protein (441 nt) |
| MGG_00585 | 11.93663794 | Up | predicted protein (354 nt) |
| MGG_14301 | 11.60269887 | Up | calpastatin (393 nt) |
| MGG_07669 | 11.58871464 | Up | predicted protein (351 nt) |
| MGG_02236 | 11.29920802 | Up | conserved hypothetical protein (366 nt) |
| MGG_09844 | 11.03617361 | Up | predicted protein (564 nt) |
| MGG_08940 | 10.69696753 | Up | predicted protein (873 nt) |
| MGG_05104 | 10.58871464 | Up | predicted protein (576 nt) |
| MGG_14376 | 10.39874369 | Up | conserved hypothetical protein (2001 nt) |
| MGG_08946 | 10.2644426 | Up | predicted protein (711 nt) |
| MGG_08352 | 9.980139578 | Up | isoflavone reductase family protein (999 nt) |
| MGG_05553 | 9.797661526 | Up | DUF895 domain membrane protein (1446 nt) |
| MGG_13037 | 9.550746785 | Up | conserved hypothetical protein (1692 nt) |
| MGG_12423 | 9.344295908 | Up | choline monooxygenase (1308 nt) |
| MGG_09972 | 9.276124405 | Up | conserved hypothetical protein (1605 nt) |
| MGG_15318 | 9.252665432 | Up | conserved hypothetical protein (1392 nt) |
| MGG_00018 | 9.022367813 | Up | integral membrane protein (1650 nt) |
| MGG_02058 | 8.965784285 | Up | mitochondrial chaperone BCS1 (1701 nt) |
| MGG_08944 | 6.118774553 | Up | predicted protein (429 nt) |
| MGG_09659 | 5.502500341 | Up | S-(hydroxymethyl)glutathione dehydrogenase (1233 nt) |
| MGG_11084 | 5.112842837 | Up | TPR domain-containing protein (2241 nt) |
| MGG_05551 | 4.584962501 | Up | conserved hypothetical protein (1521 nt) |
| MGG_15149 | 4.584962501 | Up | conserved hypothetical protein (1521 nt) |
| MGG_02067 | 4.024585638 | Up | conserved hypothetical protein (738 nt) |
| MGG_08723 | 3.855717944 | Up | conserved hypothetical protein (972 nt) |
| MGG_09645 | 3.749534268 | Up | fatty acid synthase (6381 nt) |
| MGG_08431 | 3.488286481 | Up | conserved hypothetical protein (1044 nt) |
| MGG_08950 | 3.485426827 | Up | high-affinity glucose transporter (1395 nt) |
| MGG_05747 | 3.48174178 | Up | predicted protein (426 nt) |
| MGG_14871 | 3.466699619 | Up | bifunctional molybdenum cofactor biosynthesis protein (801 nt) |
| MGG_04437 | 3.37914914 | Up | heat shock protein 78 (2421 nt) |
| MGG_12501 | 3.378511623 | Up | conserved hypothetical protein (1374 nt) |
| MGG_13924 | 3.356380304 | Up | serine palmitoyltransferase 2 (2250 nt) |
| MGG_08943 | 3.331514144 | Up | conserved hypothetical protein (957 nt) |
| MGG_05084 | 3.328187085 | Up | conserved hypothetical protein (636 nt) |
| MGG_01989 | 3.266786541 | Up | predicted protein (1665 nt) |
| MGG_07246 | 3.249359469 | Up | conserved hypothetical protein (600 nt) |
| MGG_02060 | 3.245873855 | Up | aromatic amino acid aminotransferase (1143 nt) |
| MGG_10934 | 3.177120503 | Up | predicted protein (381 nt) |
| MGG_08962 | 3.126031175 | Up | conserved hypothetical protein (546 nt) |
| MGG_14195 | 3.117512049 | Up | predicted protein (417 nt) |
| MGG_13912 | 3.10433666 | Up | conserved hypothetical protein (1482 nt) |
| MGG_09100 | 3.064822001 | Up | cutinase (645 nt) |
| MGG_13116 | 3.062735755 | Up | conserved hypothetical protein (1260 nt) |
| MGG_13923 | 3.062685268 | Up | predicted protein (582 nt) |
| MGG_09221 | 3.03562391 | Up | predicted protein (3054 nt) |
| MGG_08430 | 3.011713421 | Up | retinol dehydrogenase 12 (933 nt) |
| MGG_03395 | 2.967185849 | Up | predicted protein (471 nt) |
| MGG_06753 | 2.921840937 | Up | predicted protein (465 nt) |
| MGG_00822 | 2.888354644 | Up | predicted protein (912 nt) |
| MGG_10110 | 2.884080713 | Up | conserved hypothetical protein (441 nt) |
| MGG_07952 | 2.865316383 | Up | predicted protein (516 nt) |
| MGG_10158 | 2.857980995 | Up | QDE-2-interacting protein (1713 nt) |
| MGG_05896 | 2.815494267 | Up | predicted protein (309 nt) |
| MGG_07979 | 2.776103988 | Up | DUF563 domain-containing protein (1554 nt) |
| MGG_07953 | 2.759123954 | Up | bifunctional P-450:NADPH-P450 reductase (3258 nt) |
| MGG_08948 | 2.756784894 | Up | predicted protein (666 nt) |
| MGG_02234 | 2.748451166 | Up | conserved hypothetical protein (537 nt) |
| MGG_11445 | 2.727691395 | Up | hypothetical protein (750 nt) |
| MGG_03411 | 2.677599742 | Up | riboflavin transporter MCH5 (1443 nt) |
| MGG_03158 | 2.604862058 | Up | pumilio-family RNA binding repeat protein (2328 nt) |
| MGG_10954 | 2.597562538 | Up | isotrichodermin C-15 hydroxylase (1479 nt) |
| MGG_05132 | 2.590701406 | Up | conserved hypothetical protein (1086 nt) |
| MGG_08817 | 2.585869528 | Up | conserved hypothetical protein (357 nt) |
| MGG_02059 | 2.523561956 | Up | conserved hypothetical protein (9060 nt) |
| MGG_03983 | 2.514368617 | Up | Hsp70 nucleotide exchange factor FES1 (654 nt) |
| MGG_02350 | 2.463656204 | Up | conserved hypothetical protein (927 nt) |
| MGG_08371 | 2.452126818 | Up | predicted protein (810 nt) |
| MGG_00080 | 2.451025299 | Up | hypothetical protein (429 nt) |
| MGG_10109 | 2.438920966 | Up | conserved hypothetical protein (465 nt) |
| MGG_09138 | 2.42053125 | Up | glutathione S-transferase II (672 nt) |
| MGG_09632 | 2.396352936 | Up | conserved hypothetical protein (1044 nt) |
| MGG_07404 | 2.387023123 | Up | tripeptidyl-peptidase 1 (1806 nt) |
| MGG_09139 | 2.38669835 | Up | laccase-1 (1794 nt) |
| MGG_06759 | 2.364189581 | Up | heat shock protein 80 (2109 nt) |
| MGG_10271 | 2.342392197 | Up | conserved hypothetical protein (3960 nt) |
| MGG_05554 | 2.329422631 | Up | conserved hypothetical protein (1401 nt) |
| MGG_02436 | 2.296338285 | Up | conserved hypothetical protein (459 nt) |
| MGG_10415 | 2.270089163 | Up | predicted protein (2403 nt) |
| MGG_05564 | 2.254813899 | Up | predicted protein (753 nt) |
| MGG_06459 | 2.200152894 | Up | heat shock protein HSP98 (2781 nt) |
| MGG_09474 | 2.179085001 | Up | hypothetical protein (504 nt) |
| MGG_09241 | 2.178803153 | Up | predicted protein (801 nt) |
| MGG_02235 | 2.174370906 | Up | oxidoreductase (996 nt) |
| MGG_07949 | 2.170357399 | Up | conserved hypothetical protein (879 nt) |
| MGG_10108 | 2.167556766 | Up | conserved hypothetical protein (789 nt) |
| MGG_08947 | 2.165337732 | Up | cytochrome P450 (1626 nt) |
| MGG_03115 | 2.145452682 | Up | predicted protein (1401 nt) |
| MGG_05819 | 2.131244533 | Up | conserved hypothetical protein (690 nt) |
| MGG_10205 | 2.111202562 | Up | isoleucyl-tRNA synthetase (1269 nt) |
| MGG_09381 | 2.104530583 | Up | conserved hypothetical protein (474 nt) |
| MGG_03495 | 2.096526924 | Up | predicted protein (528 nt) |
| MGG_05824 | 2.068901305 | Up | UDP-glucuronosyl/UDP-glucosyltransferase (1299 nt) |
| MGG_06958 | 2.06641867 | Up | hsp70-like protein (1956 nt) |
| MGG_07954 | 2.03876957 | Up | epoxide hydrolase 2 (1017 nt) |
| MGG_11021 | 2.02307246 | Up | conserved hypothetical protein (2061 nt) |
| MGG_02372 | 2.018018704 | Up | hypothetical protein (729 nt) |
| MGG_08424 | 2.0138058 | Up | endo-1,4-beta-xylanase I (696 nt) |
| MGG_07615 | 2.006853671 | Up | protoporphyrinogen oxidase (693 nt) |
| MGG_06298 | 1.989920444 | Up | hypothetical protein (1176 nt) |
| MGG_07646 | 1.983511877 | Up | alpha-glucuronidase (2592 nt) |
| MGG_10912 | 1.959738155 | Up | mycocerosic acid synthase (7641 nt) |
| MGG_07219 | 1.949373927 | Up | conidial yellow pigment biosynthesis polyketide synthase (6528 nt) |
| MGG_14351 | 1.94240208 | Up | glycosyltransferase (1269 nt) |
| MGG_03471 | 1.939458458 | Up | predicted protein (1176 nt) |
| MGG_14703 | 1.937959253 | Up | conserved hypothetical protein (387 nt) |
| MGG_14340 | 1.934604368 | Up | predicted protein (324 nt) |
| MGG_14375 | 1.902364997 | Up | conserved hypothetical protein (609 nt) |
| MGG_15049 | 1.871675903 | Up | predicted protein (1944 nt) |
| MGG_02267 | 1.866733469 | Up | predicted protein (1044 nt) |
| MGG_10683 | 1.849665727 | Up | predicted protein (1479 nt) |
| MGG_03238 | 1.848445436 | Up | zinc finger protein ZPR1 (1533 nt) |
| MGG_02095 | 1.839416707 | Up | conserved hypothetical protein (996 nt) |
| MGG_08378 | 1.824913293 | Up | cytochrome P450 oxidoreductase GliF (1605 nt) |
| MGG_04511 | 1.821029859 | Up | MFS transporter (1461 nt) |
| MGG_07946 | 1.811554911 | Up | conserved hypothetical protein (1494 nt) |
| MGG_12697 | 1.798601538 | Up | pheromone-regulated membrane protein 10 (2973 nt) |
| MGG_05505 | 1.795986108 | Up | autophagy protein (Atg22) (1704 nt) |
| MGG_01410 | 1.791820579 | Up | glutathione S-transferase Gst3 (1035 nt) |
| MGG_02043 | 1.777192952 | Up | BTB/POZ domain-containing protein (510 nt) |
| MGG_14675 | 1.772956761 | Up | hypothetical protein (180 nt) |
| MGG_06616 | 1.762500686 | Up | high-affinity methionine permease (1833 nt) |
| MGG_11754 | 1.757836032 | Up | heavy metal tolerance protein (2163 nt) |
| MGG_02096 | 1.757618704 | Up | glycoside hydrolase (2478 nt) |
| MGG_14523 | 1.75482247 | Up | hypothetical protein (366 nt) |
| MGG_02002 | 1.744835012 | Up | conserved hypothetical protein (1281 nt) |
| MGG_02069 | 1.739398362 | Up | glyoxalase/bleomycin resistance protein/dioxygenase (396 nt) |
| MGG_10573 | 1.729422654 | Up | predicted protein (1473 nt) |
| MGG_06156 | 1.727406766 | Up | kelch repeat protein (2379 nt) |
| MGG_14621 | 1.724556807 | Up | conserved hypothetical protein (279 nt) |
| MGG_15465 | 1.720557962 | Up | hypothetical protein (729 nt) |
| MGG_11336 | 1.703898264 | Up | conserved hypothetical protein (774 nt) |
| MGG_07608 | 1.696188456 | Up | predicted protein (375 nt) |
| MGG_07216 | 1.691742463 | Up | versicolorin reductase (825 nt) |
| MGG_10921 | 1.684958291 | Up | conserved hypothetical protein (201 nt) |
| MGG_05515 | 1.669026766 | Up | predicted protein (588 nt) |
| MGG_08180 | 1.665397624 | Up | conserved hypothetical protein (1125 nt) |
| MGG_13764 | 1.664251411 | Up | bilirubin oxidase (1890 nt) |
| MGG_13110 | 1.662130361 | Up | 4-hydroxyacetophenone monooxygenase (1914 nt) |
| MGG_08989 | 1.660472735 | Up | conserved hypothetical protein (816 nt) |
| MGG_00681 | 1.647698256 | Up | conserved hypothetical protein (1215 nt) |
| MGG_15364 | 1.641546029 | Up | predicted protein (537 nt) |
| MGG_09998 | 1.637095305 | Up | hypothetical protein (282 nt) |
| MGG_09647 | 1.633461018 | Up | 2-succinylbenzoate-CoA ligase (1506 nt) |
| MGG_13019 | 1.631716269 | Up | predicted protein (690 nt) |
| MGG_11680 | 1.620690205 | Up | AAA family ATPase (2112 nt) |
| MGG_10327 | 1.616246789 | Up | conserved hypothetical protein (2121 nt) |
| MGG_15048 | 1.612022843 | Up | NAD-dependent deacetylase sirtuin-5 (1074 nt) |
| MGG_08949 | 1.604478457 | Up | hypothetical protein (1233 nt) |
| MGG_00791 | 1.603109847 | Up | Lactamase_B domain containing protein (1077 nt) |
| MGG_05046 | 1.579829654 | Up | adenine phosphoribosyltransferase (3519 nt) |
| MGG_03034 | 1.574139531 | Up | conserved hypothetical protein (1581 nt) |
| MGG_06234 | 1.567215347 | Up | predicted protein (426 nt) |
| MGG_04627 | 1.566815154 | Up | conserved hypothetical protein (2787 nt) |
| MGG_00287 | 1.56664049 | Up | predicted protein (528 nt) |
| MGG_08519 | 1.565654069 | Up | aflatoxin B1 aldehyde reductase member 3 (1002 nt) |
| MGG_12714 | 1.563004767 | Up | conserved hypothetical protein (930 nt) |
| MGG_14872 | 1.551770974 | Up | calpain-9 (2604 nt) |
| MGG_08941 | 1.543130572 | Up | predicted protein (615 nt) |
| MGG_13562 | 1.541985428 | Up | conserved hypothetical protein (1593 nt) |
| MGG_01454 | 1.541813265 | Up | conserved hypothetical protein (720 nt) |
| MGG_01364 | 1.531668886 | Up | conserved hypothetical protein (2925 nt) |
| MGG_08980 | 1.528536579 | Up | stress-induced-phosphoprotein 1 (1755 nt) |
| MGG_05418 | 1.526747887 | Up | predicted protein (852 nt) |
| MGG_11991 | 1.516744272 | Up | predicted protein (252 nt) |
| MGG_15420 | 1.515463451 | Up | predicted protein (498 nt) |
| MGG_07766 | 1.514203204 | Up | predicted protein (372 nt) |
| MGG_03396 | 1.501631699 | Up | conserved hypothetical protein (2049 nt) |
| MGG_04736 | 1.500318333 | Up | conserved hypothetical protein (912 nt) |
| MGG_15188 | 1.496902147 | Up | conserved hypothetical protein (567 nt) |
| MGG_13118 | 1.49523234 | Up | predicted protein (555 nt) |
| MGG_02065 | 1.492089159 | Up | kinesin light chain (2622 nt) |
| MGG_05968 | 1.483038711 | Up | endoribonuclease L-PSP (411 nt) |
| MGG_02977 | 1.479542529 | Up | predicted protein (1809 nt) |
| MGG_02854 | 1.47566256 | Up | conserved hypothetical protein (942 nt) |
| MGG_05555 | 1.474172623 | Up | xenobiotic compound monooxygenase (1476 nt) |
| MGG_08726 | 1.47279708 | Up | hypothetical protein (588 nt) |
| MGG_00809 | 1.472177608 | Up | predicted protein (1770 nt) |
| MGG_06957 | 1.449286928 | Up | dienelactone hydrolase family protein (852 nt) |
| MGG_11779 | 1.447112965 | Up | conserved hypothetical protein (5598 nt) |
| MGG_07402 | 1.436863862 | Up | conserved hypothetical protein (465 nt) |
| MGG_13253 | 1.434425855 | Up | choline dehydrogenase (1977 nt) |
| MGG_08373 | 1.427917291 | Up | conserved hypothetical protein (552 nt) |
| MGG_01439 | 1.425133377 | Up | inorganic phosphate transporter PHO84 (2352 nt) |
| MGG_13839 | 1.42203958 | Up | predicted protein (408 nt) |
| MGG_14692 | 1.420240156 | Up | conserved hypothetical protein (1233 nt) |
| MGG_00832 | 1.402635441 | Up | cytochrome P450 (1605 nt) |
| MGG_15168 | 1.402503146 | Up | predicted protein (1179 nt) |
| MGG_15151 | 1.40118504 | Up | hypothetical protein (900 nt) |
| MGG_06779 | 1.394108478 | Up | NADP-dependent mannitol dehydrogenase (816 nt) |
| MGG_08486 | 1.385724603 | Up | beta-lactamase family protein (1698 nt) |
| MGG_09019 | 1.3852473 | Up | secretory phospholipase A2 (561 nt) |
| MGG_03585 | 1.381283373 | Up | predicted protein (420 nt) |
| MGG_08429 | 1.381090167 | Up | serin endopeptidas (2595 nt) |
| MGG_04258 | 1.375077953 | Up | predicted protein (423 nt) |
| MGG_08677 | 1.358748368 | Up | predicted protein (1065 nt) |
| MGG_02252 | 1.352329115 | Up | tetrahydroxynaphthalene reductase (852 nt) |
| MGG_07078 | 1.347104217 | Up | DUF775 domain-containing protein (609 nt) |
| MGG_08434 | 1.34533795 | Up | conserved hypothetical protein (3564 nt) |
| MGG_15466 | 1.339875928 | Up | conserved hypothetical protein (327 nt) |
| MGG_15150 | 1.339875928 | Up | conserved hypothetical protein (327 nt) |
| MGG_02056 | 1.337755578 | Up | predicted protein (1062 nt) |
| MGG_05590 | 1.337107236 | Up | zeaxanthin epoxidase (1197 nt) |
| MGG_09131 | 1.334601925 | Up | alpha/beta hydrolase (861 nt) |
| MGG_09168 | 1.330645312 | Up | predicted protein (732 nt) |
| MGG_00156 | 1.330364932 | Up | conserved hypothetical protein (966 nt) |
| MGG_10272 | 1.329656431 | Up | conserved hypothetical protein (864 nt) |
| MGG_00963 | 1.327804661 | Up | predicted protein (2850 nt) |
| MGG_02648 | 1.327556301 | Up | interferon-induced GTP-binding protein Mx (2259 nt) |
| MGG_07607 | 1.303355989 | Up | conserved hypothetical protein (477 nt) |
| MGG_10207 | 1.295674855 | Up | predicted protein (1449 nt) |
| MGG_05574 | 1.292180751 | Up | conserved hypothetical protein (1539 nt) |
| MGG_05419 | 1.290864083 | Up | predicted protein (2484 nt) |
| MGG_08451 | 1.288093497 | Up | predicted protein (333 nt) |
| MGG_08029 | 1.281729267 | Up | conserved hypothetical protein (942 nt) |
| MGG_11682 | 1.281189416 | Up | phenylacetone monooxygenase (1674 nt) |
| MGG_10450 | 1.276331228 | Up | conserved hypothetical protein (3099 nt) |
| MGG_00399 | 1.268384642 | Up | DUF833 domain-containing protein (1191 nt) |
| MGG_07146 | 1.262639835 | Up | glycosyl hydrolase (2286 nt) |
| MGG_00282 | 1.258104518 | Up | conserved hypothetical protein (2610 nt) |
| MGG_08104 | 1.254630144 | Up | 41 kDa peptidyl-prolyl cis-trans isomerase (1131 nt) |
| MGG_01951 | 1.25334101 | Up | 4-coumarate-CoA ligase 2 (2010 nt) |
| MGG_05012 | 1.248761682 | Up | predicted protein (453 nt) |
| MGG_00814 | 1.246729595 | Up | Hsp90 co-chaperone AHA1 (984 nt) |
| MGG_01420 | 1.239727032 | Up | conserved hypothetical protein (1524 nt) |
| MGG_11409 | 1.239290946 | Up | predicted protein (363 nt) |
| MGG_15105 | 1.235134723 | Up | predicted protein (795 nt) |
| MGG_07367 | 1.22804954 | Up | conserved hypothetical protein (831 nt) |
| MGG_10911 | 1.22795486 | Up | cupin domain-containing protein (597 nt) |
| MGG_08158 | 1.217117479 | Up | predicted protein (993 nt) |
| MGG_05978 | 1.216910356 | Up | conserved hypothetical protein (564 nt) |
| MGG_05913 | 1.213091603 | Up | hypothetical protein (657 nt) |
| MGG_10716 | 1.196161698 | Up | conserved hypothetical protein (540 nt) |
| MGG_04757 | 1.193739826 | Up | conserved hypothetical protein (1485 nt) |
| MGG_05059 | 1.193074321 | Up | scytalone dehydratase (519 nt) |
| MGG_04346 | 1.189968987 | Up | sterol 24-C-methyltransferase (1173 nt) |
| MGG_15230 | 1.186570067 | Up | hypothetical protein (207 nt) |
| MGG_05611 | 1.181656367 | Up | hypothetical protein (243 nt) |
| MGG_09384 | 1.160059648 | Up | conserved hypothetical protein (1542 nt) |
| MGG_05486 | 1.158382958 | Up | conserved hypothetical protein (1053 nt) |
| MGG_08990 | 1.15218757 | Up | predicted protein (882 nt) |
| MGG_07571 | 1.142006602 | Up | LysM domain-containing protein (429 nt) |
| MGG_07227 | 1.141693504 | Up | hypothetical protein (468 nt) |
| MGG_06155 | 1.137106032 | Up | hypothetical protein (1782 nt) |
| MGG_00834 | 1.136488996 | Up | hypothetical protein (987 nt) |
| MGG_03822 | 1.128882769 | Up | peptidase family T4 (1185 nt) |
| MGG_14857 | 1.126377286 | Up | conserved hypothetical protein (1050 nt) |
| MGG_15342 | 1.12210343 | Up | predicted protein (1407 nt) |
| MGG_08459 | 1.118055762 | Up | ent-kaurene oxidase (1422 nt) |
| MGG_02114 | 1.117237675 | Up | conserved hypothetical protein (2439 nt) |
| MGG_02064 | 1.115477217 | Up | conserved hypothetical protein (1167 nt) |
| MGG_02105 | 1.112563575 | Up | conserved hypothetical protein (915 nt) |
| MGG_05985 | 1.110182918 | Up | conserved hypothetical protein (2136 nt) |
| MGG_03005 | 1.109080361 | Up | conserved hypothetical protein (1332 nt) |
| MGG_08007 | 1.107108893 | Up | conserved hypothetical protein (762 nt) |
| MGG_00046 | 1.097054684 | Up | conserved hypothetical protein (2739 nt) |
| MGG_11608 | 1.096844325 | Up | laccase-2 (1794 nt) |
| MGG_15462 | 1.095964604 | Up | hypothetical protein (222 nt) |
| MGG_03442 | 1.092775669 | Up | conserved hypothetical protein (702 nt) |
| MGG_01596 | 1.088810859 | Up | DNA damage response protein kinase DUN1 (2025 nt) |
| MGG_14651 | 1.08767481 | Up | conserved hypothetical protein (696 nt) |
| MGG_04242 | 1.081893423 | Up | bax Inhibitor family protein (786 nt) |
| MGG_00121 | 1.08161991 | Up | conserved hypothetical protein (537 nt) |
| MGG_00292 | 1.079165336 | Up | conserved hypothetical protein (612 nt) |
| MGG_09664 | 1.075597619 | Up | beta-mannosidase (2739 nt) |
| MGG_06131 | 1.073159746 | Up | conserved hypothetical protein (720 nt) |
| MGG_08612 | 1.072360223 | Up | predicted protein (1851 nt) |
| MGG_05940 | 1.070943772 | Up | short chain dehydrogenase (780 nt) |
| MGG_07577 | 1.070389328 | Up | conserved hypothetical protein (1821 nt) |
| MGG_02061 | 1.067002855 | Up | conserved hypothetical protein (2025 nt) |
| MGG_07315 | 1.066011253 | Up | predicted protein (411 nt) |
| MGG_00078 | 1.063578796 | Up | conserved hypothetical protein (1188 nt) |
| MGG_10613 | 1.054447784 | Up | retinol dehydrogenase 11 (975 nt) |
| MGG_08142 | 1.052911531 | Up | conserved hypothetical protein (1305 nt) |
| MGG_05567 | 1.05288558 | Up | conserved hypothetical protein (573 nt) |
| MGG_06610 | 1.051720297 | Up | lipase (1203 nt) |
| MGG_03098 | 1.049687124 | Up | conserved hypothetical protein (984 nt) |
| MGG_09328 | 1.048429433 | Up | conserved hypothetical protein (1113 nt) |
| MGG_07818 | 1.043795118 | Up | conserved hypothetical protein (2484 nt) |
| MGG_11355 | 1.041169189 | Up | predicted protein (1995 nt) |
| MGG_01489 | 1.04029647 | Up | conserved hypothetical protein (2940 nt) |
| MGG_14061 | 1.039647809 | Up | oxalate decarboxylase oxdC (1503 nt) |
| MGG_10896 | 1.038395379 | Up | allantoate permease (1578 nt) |
| MGG_05656 | 1.035868248 | Up | ankyrin repeat protein (5400 nt) |
| MGG_00591 | 1.034995421 | Up | predicted protein (1677 nt) |
| MGG_04569 | 1.029318937 | Up | NADPH dehydrogenase (1329 nt) |
| MGG_00738 | 1.025321666 | Up | zinc-binding alcohol dehydrogenase domain-containing protein 2 (1020 nt) |
| MGG_13360 | 1.021398037 | Up | conserved hypothetical protein (1653 nt) |
| MGG_02355 | 1.018265026 | Up | predicted protein (687 nt) |
| MGG_10097 | 1.017080484 | Up | intracellular hyphae protein 1 (489 nt) |
| MGG_08020 | 1.014080909 | Up | endoglucanase-4 (729 nt) |
| MGG_13119 | 1.009407453 | Up | predicted protein (1401 nt) |
| MGG_12222 | 1.009179427 | Up | F-box domain-containing protein (1383 nt) |
| MGG_02445 | 1.008878151 | Up | predicted protein (801 nt) |
| MGG_05045 | 1.008787304 | Up | GTP cyclohydrolase I (777 nt) |
| MGG_10893 | 1.00877021 | Up | conserved hypothetical protein (1548 nt) |
| MGG_01692 | 1.008721271 | Up | conserved hypothetical protein (1206 nt) |
| MGG_05880 | 1.00264149 | Up | glycerol uptake facilitator protein (1173 nt) |
| MGG_14668 | -12.80976813 | Down | hypothetical protein (228 nt) |
| MGG_11005 | -12.36632221 | Down | conserved hypothetical protein (1368 nt) |
| MGG_14997 | -12.14210706 | Down | hypothetical protein (219 nt) |
| MGG_14426 | -11.69696753 | Down | conserved hypothetical protein (609 nt) |
| MGG_14841 | -11.62113611 | Down | predicted protein (342 nt) |
| MGG_07726 | -11.50183718 | Down | conserved hypothetical protein (369 nt) |
| MGG_15120 | -11.15481811 | Down | hypothetical protein (399 nt) |
| MGG_09144 | -10.96578428 | Down | predicted protein (450 nt) |
| MGG_14926 | -10.92925841 | Down | predicted protein (531 nt) |
| MGG_10040 | -10.12928302 | Down | ferulic acid esterase A (1023 nt) |
| MGG_01975 | -9.252665432 | Down | predicted protein (2526 nt) |
| MGG_09490 | -8.965784285 | Down | predicted protein (1701 nt) |
| MGG_15409 | -4.635588574 | Down | predicted protein (432 nt) |
| MGG_03044 | -4.310142181 | Down | conserved hypothetical protein (1101 nt) |
| MGG_02942 | -4.161723188 | Down | conserved hypothetical protein (1749 nt) |
| MGG_07807 | -4.138216142 | Down | conserved hypothetical protein (648 nt) |
| MGG_09436 | -4.034269902 | Down | conserved hypothetical protein (303 nt) |
| MGG_09879 | -3.992938336 | Down | conserved hypothetical protein (438 nt) |
| MGG_03349 | -3.849175098 | Down | auxin Efflux Carrier superfamily (1314 nt) |
| MGG_00766 | -3.619372938 | Down | conserved hypothetical protein (558 nt) |
| MGG_00824 | -3.572889668 | Down | phenylacetone monooxygenase (1890 nt) |
| MGG_10751 | -3.440572591 | Down | peroxisomal copper amine oxidase (2025 nt) |
| MGG_01298 | -3.423211431 | Down | malic acid transport protein (1323 nt) |
| MGG_07857 | -3.413850584 | Down | conserved hypothetical protein (1125 nt) |
| MGG_10650 | -3.408267701 | Down | hypothetical protein (291 nt) |
| MGG_03825 | -3.273018494 | Down | isotrichodermin C-15 hydroxylase (1617 nt) |
| MGG_05250 | -3.245205238 | Down | conserved hypothetical protein (2520 nt) |
| MGG_10593 | -3.2410081 | Down | predicted protein (456 nt) |
| MGG_14237 | -3.214407437 | Down | predicted protein (288 nt) |
| MGG_09994 | -3.174512271 | Down | lipid phosphate phosphatase 1 (1140 nt) |
| MGG_03329 | -3.169891506 | Down | small heat shock protein (633 nt) |
| MGG_05165 | -3.15264701 | Down | conserved hypothetical protein (1533 nt) |
| MGG_02290 | -3.141142101 | Down | conserved hypothetical protein (858 nt) |
| MGG_08416 | -3.070389328 | Down | lipase 1 (1674 nt) |
| MGG_01391 | -3.070166018 | Down | ent-kaurene oxidase (1569 nt) |
| MGG_07713 | -3.06833713 | Down | predicted protein (453 nt) |
| MGG_09418 | -3.02188266 | Down | conserved hypothetical protein (429 nt) |
| MGG_11915 | -2.901295133 | Down | predicted protein (393 nt) |
| MGG_08971 | -2.886958693 | Down | conserved hypothetical protein (573 nt) |
| MGG_08326 | -2.881995905 | Down | conserved hypothetical protein (1416 nt) |
| MGG_10322 | -2.87570589 | Down | cortical patch protein (765 nt) |
| MGG_02336 | -2.814444347 | Down | isoflavone reductase family protein (981 nt) |
| MGG_09134 | -2.765762647 | Down | hypothetical protein (357 nt) |
| MGG_04171 | -2.721732314 | Down | predicted protein (1050 nt) |
| MGG_14581 | -2.718799594 | Down | hypothetical protein (744 nt) |
| MGG_07348 | -2.697353738 | Down | predicted protein (420 nt) |
| MGG_04075 | -2.688913465 | Down | lambda-crystallin (993 nt) Note(s): contains exonic Ns |
| MGG_10713 | -2.660707862 | Down | conserved hypothetical protein (813 nt) |
| MGG_02329 | -2.649306721 | Down | isotrichodermin C-15 hydroxylase (1632 nt) |
| MGG_00733 | -2.605714662 | Down | conserved hypothetical protein (1026 nt) |
| MGG_07354 | -2.579487371 | Down | glutathionylspermidine synthase (1296 nt) |
| MGG_12800 | -2.531615468 | Down | conserved hypothetical protein (1257 nt) |
| MGG_01152 | -2.496200352 | Down | conserved hypothetical protein (549 nt) |
| MGG_07871 | -2.493092449 | Down | conserved hypothetical protein (726 nt) |
| MGG_08498 | -2.492768803 | Down | isotrichodermin C-15 hydroxylase (1506 nt) |
| MGG_07697 | -2.470862199 | Down | superoxide dismutase (642 nt) |
| MGG_07556 | -2.435386145 | Down | conserved hypothetical protein (672 nt) |
| MGG_04699 | -2.425911543 | Down | C2H2 finger domain-containing protein FlbC (1179 nt) |
| MGG_12865 | -2.412767326 | Down | conserved hypothetical protein (1344 nt) |
| MGG_06337 | -2.409390936 | Down | conserved hypothetical protein (3441 nt) |
| MGG_00040 | -2.400827014 | Down | high-affinity glucose transporter ght2 (1713 nt) |
| MGG_11876 | -2.393623513 | Down | sugar transporter STL1 (1722 nt) |
| MGG_01811 | -2.370837695 | Down | conserved hypothetical protein (3165 nt) |
| MGG_01368 | -2.345597001 | Down | conserved hypothetical protein (714 nt) |
| MGG_10714 | -2.315031003 | Down | cyanide hydratase (1071 nt) |
| MGG_03739 | -2.30256277 | Down | predicted protein (2823 nt) |
| MGG_03977 | -2.298702913 | Down | hypothetical protein (1389 nt) |
| MGG_01369 | -2.295885623 | Down | alpha/beta hydrolase fold-3 domain-containing protein (1044 nt) |
| MGG_10429 | -2.277318632 | Down | conserved hypothetical protein (963 nt) |
| MGG_06513 | -2.271567134 | Down | hypothetical protein (369 nt) |
| MGG_05804 | -2.269857074 | Down | phospholipase D p1 (2574 nt) |
| MGG_13655 | -2.268229568 | Down | conserved hypothetical protein (1416 nt) |
| MGG_09445 | -2.266500219 | Down | conserved hypothetical protein (1437 nt) |
| MGG_07088 | -2.257691984 | Down | hypothetical protein (315 nt) |
| MGG_12086 | -2.254241287 | Down | conserved hypothetical protein (759 nt) |
| MGG_05798 | -2.248888349 | Down | conserved hypothetical protein (651 nt) |
| MGG_09473 | -2.24858331 | Down | hypothetical protein (354 nt) |
| MGG_09000 | -2.230954435 | Down | protein kinase domain-containing protein (2160 nt) |
| MGG_10268 | -2.209640685 | Down | MAP kinase kinase PBS2 (2058 nt) |
| MGG_10498 | -2.201633861 | Down | conserved hypothetical protein (1044 nt) |
| MGG_08405 | -2.190222416 | Down | retinol dehydrogenase 13 (969 nt) |
| MGG_06545 | -2.168411949 | Down | conserved hypothetical protein (2358 nt) |
| MGG_08439 | -2.161473337 | Down | predicted protein (846 nt) |
| MGG_13198 | -2.159984689 | Down | predicted protein (270 nt) |
| MGG_09261 | -2.13827611 | Down | hypothetical protein (969 nt) |
| MGG_02340 | -2.118644496 | Down | cell wall glycosyl hydrolase YteR (1218 nt) |
| MGG_04254 | -2.107640723 | Down | conserved hypothetical protein (621 nt) |
| MGG_01431 | -2.102966579 | Down | predicted protein (462 nt) |
| MGG_03469 | -2.083854051 | Down | predicted protein (714 nt) |
| MGG_02943 | -2.079420312 | Down | predicted protein (1011 nt) |
| MGG_15283 | -2.073777926 | Down | NAD dependent epimerase/dehydratase family protein (804 nt) |
| MGG_01542 | -2.072756342 | Down | endo-1,4-beta-xylanase (996 nt) |
| MGG_00127 | -2.070866331 | Down | saccharopine dehydrogenase (1290 nt) |
| MGG_05719 | -2.05779793 | Down | 30 kDa heat shock protein (723 nt) |
| MGG_09851 | -2.05288558 | Down | predicted protein (1029 nt) |
| MGG_08277 | -2.051977291 | Down | predicted protein (867 nt) |
| MGG_12366 | -2.051002903 | Down | hypothetical protein (162 nt) |
| MGG_07356 | -2.050626073 | Down | predicted protein (1701 nt) |
| MGG_02850 | -2.044818504 | Down | conserved hypothetical protein (846 nt) |
| MGG_10503 | -2.038908043 | Down | mannitol-1-phosphate 5-dehydrogenase (1203 nt) |
| MGG_21009 | -2.035128223 | Down | Magnaporthe grisea 70-15 mitochondria cytochrome c oxidase subunit I cox1 (1599 nt) |
| MGG_02709 | -2.016440916 | Down | conserved hypothetical protein (465 nt) |
| MGG_10456 | -2.014882446 | Down | hypothetical protein (450 nt) |
| MGG_15428 | -2.012748493 | Down | para-nitrobenzyl esterase (1674 nt) |
| MGG_03764 | -2 | Down | salicylate hydroxylase (1395 nt) |
| MGG_08059 | -1.99175344 | Down | conserved hypothetical protein (615 nt) |
| MGG_15427 | -1.991282783 | Down | predicted protein (705 nt) |
| MGG_10497 | -1.989731665 | Down | blue light-inducible protein Bli-3 (627 nt) |
| MGG_02803 | -1.972238488 | Down | hypothetical protein (207 nt) |
| MGG_04251 | -1.96800463 | Down | phosphate transporter family protein (1782 nt) |
| MGG_10107 | -1.950287788 | Down | caleosin domain-containing protein (750 nt) |
| MGG_04561 | -1.942052651 | Down | DNA repair family protein (912 nt) |
| MGG_12471 | -1.921390165 | Down | predicted protein (735 nt) |
| MGG_02302 | -1.920565533 | Down | HhH-GPD family base excision DNA repair protein (1128 nt) |
| MGG_09001 | -1.915662644 | Down | hypothetical protein (474 nt) |
| MGG_00114 | -1.914793825 | Down | conserved hypothetical protein (594 nt) |
| MGG_00677 | -1.902180799 | Down | endoglucanase-1 (774 nt) |
| MGG_06865 | -1.901041095 | Down | hypothetical protein (450 nt) |
| MGG_00993 | -1.89630787 | Down | vacuolar iron transporter Ccc1 (1014 nt) |
| MGG_03643 | -1.885795281 | Down | conserved hypothetical protein (3816 nt) |
| MGG_15068 | -1.872299651 | Down | hypothetical protein (399 nt) |
| MGG_11813 | -1.864915077 | Down | predicted protein (738 nt) |
| MGG_00126 | -1.862496476 | Down | conserved hypothetical protein (2886 nt) |
| MGG_01373 | -1.846069502 | Down | arabinose-proton symporter (1728 nt) |
| MGG_14292 | -1.812914447 | Down | aminopeptidase Y (1440 nt) |
| MGG_03341 | -1.801095932 | Down | galactose-proton symporter (1716 nt) |
| MGG_07806 | -1.800230488 | Down | conserved hypothetical protein (1230 nt) |
| MGG_04203 | -1.799556551 | Down | 4-coumarate-CoA ligase 1 (1671 nt) |
| MGG_10730 | -1.793549123 | Down | calcium-transporting ATPase 3 (3285 nt) |
| MGG_01544 | -1.788997336 | Down | pisatin demethylase (1464 nt) |
| MGG_09852 | -1.788893386 | Down | sugar transporter STL1 (1707 nt) |
| MGG_08883 | -1.771558534 | Down | conserved hypothetical protein (2241 nt) |
| MGG_03987 | -1.767305023 | Down | predicted protein (1773 nt) |
| MGG_14146 | -1.764469109 | Down | hypothetical protein (384 nt) |
| MGG_09828 | -1.763810581 | Down | vacuolar conductance protein (1959 nt) |
| MGG_10515 | -1.745603911 | Down | conserved hypothetical protein (966 nt) |
| MGG_04165 | -1.74159068 | Down | conserved hypothetical protein (3669 nt) |
| MGG_02752 | -1.734749061 | Down | hypothetical protein (1071 nt) |
| MGG_03575 | -1.73033967 | Down | conserved hypothetical protein (372 nt) |
| MGG_02893 | -1.727525065 | Down | conserved hypothetical protein (468 nt) |
| MGG_10147 | -1.727109398 | Down | multidrug resistance protein fnx1 (1719 nt) |
| MGG_02641 | -1.724508709 | Down | conserved hypothetical protein (873 nt) |
| MGG_14591 | -1.715023041 | Down | cytochrome P450 3A24 (1617 nt) |
| MGG_05358 | -1.714428705 | Down | LYR family protein (267 nt) |
| MGG_06567 | -1.708110731 | Down | conserved hypothetical protein (996 nt) |
| MGG_09323 | -1.703160095 | Down | phenylacetone monooxygenase (1944 nt) |
| MGG_08581 | -1.701809149 | Down | conserved hypothetical protein (1278 nt) |
| MGG_08770 | -1.700439718 | Down | conserved hypothetical protein (1539 nt) |
| MGG_09260 | -1.697890657 | Down | integral membrane protein (495 nt) |
| MGG_05750 | -1.695030732 | Down | conserved hypothetical protein (933 nt) |
| MGG_13798 | -1.686291456 | Down | predicted protein (915 nt) |
| MGG_07966 | -1.684741955 | Down | phosphate transporter family protein (1767 nt) |
| MGG_09818 | -1.683468858 | Down | conserved hypothetical protein (1359 nt) |
| MGG_02929 | -1.675273272 | Down | phosphotransferase enzyme family protein (789 nt) |
| MGG_10359 | -1.673771768 | Down | conserved hypothetical protein (1248 nt) |
| MGG_09116 | -1.672705395 | Down | xylulose-5-phosphate/fructose-6-phosphate phosphoketolase (2433 nt) |
| MGG_04257 | -1.672160943 | Down | secretory lipase family protein (1269 nt) |
| MGG_02117 | -1.671216173 | Down | conserved hypothetical protein (462 nt) |
| MGG_07890 | -1.667424661 | Down | aldehyde dehydrogenase 3I1 (1482 nt) |
| MGG_05164 | -1.665356281 | Down | conserved hypothetical protein (1704 nt) |
| MGG_02246 | -1.659294698 | Down | conserved hypothetical protein (258 nt) |
| MGG_02332 | -1.661898167 | Down | conserved hypothetical protein (810 nt) |
| MGG_01376 | -1.657020649 | Down | tyrosine-protein phosphatase 1 (3420 nt) |
| MGG_01945 | -1.653849412 | Down | predicted protein (729 nt) |
| MGG_03336 | -1.651154546 | Down | LEA domain-containing protein (405 nt) |
| MGG_08994 | -1.649813645 | Down | conserved hypothetical protein (726 nt) |
| MGG_10973 | -1.647520814 | Down | conserved hypothetical protein (570 nt) |
| MGG_10422 | -1.646850887 | Down | C6 transcription factor OefC (2112 nt) |
| MGG_09985 | -1.628174568 | Down | hypothetical protein (1347 nt) |
| MGG_14578 | -1.620712526 | Down | predicted protein (855 nt) |
| MGG_04628 | -1.615245068 | Down | cytochrome P450 51 (1548 nt) |
| MGG_07816 | -1.607682577 | Down | predicted protein (486 nt) |
| MGG_13428 | -1.605463615 | Down | mitochondrial phosphate carrier protein (945 nt) |
| MGG_12530 | -1.59724083 | Down | histidine kinase G7 (3768 nt) |
| MGG_01798 | -1.587742596 | Down | conserved hypothetical protein (906 nt) |
| MGG_09351 | -1.580096746 | Down | aspergillopepsin-F (1293 nt) |
| MGG_12664 | -1.579226131 | Down | conserved hypothetical protein (777 nt) |
| MGG_06181 | -1.578735926 | Down | predicted protein (1539 nt) |
| MGG_03246 | -1.5776577 | Down | conserved hypothetical protein (456 nt) |
| MGG_01463 | -1.576467707 | Down | phosphate metabolism protein 7 (2682 nt) |
| MGG_10092 | -1.574527048 | Down | hypothetical protein (1086 nt) |
| MGG_09857 | -1.574236094 | Down | sorbitol dehydrogenase (1116 nt) |
| MGG_00097 | -1.569986896 | Down | conserved hypothetical protein (972 nt) |
| MGG_01260 | -1.566081696 | Down | serine/threonine-protein kinase psk1 (1770 nt) |
| MGG_08547 | -1.55844129 | Down | serine/threonine-protein kinase srk1 (1872 nt) |
| MGG_09954 | -1.557777671 | Down | conserved hypothetical protein (3147 nt) |
| MGG_08552 | -1.555690119 | Down | hypothetical protein (351 nt) |
| MGG_10394 | -1.553362167 | Down | conserved hypothetical protein (618 nt) |
| MGG_10053 | -1.552972386 | Down | conserved hypothetical protein (867 nt) |
| MGG_15228 | -1.538419915 | Down | predicted protein (354 nt) |
| MGG_05897 | -1.535135392 | Down | predicted protein (462 nt) |
| MGG_06956 | -1.531940639 | Down | predicted protein (636 nt) |
| MGG_05988 | -1.5261405 | Down | lipid phosphate phosphatase 2 (1239 nt) |
| MGG_05993 | -1.524431255 | Down | conserved hypothetical protein (2223 nt) |
| MGG_00830 | -1.519972879 | Down | conserved hypothetical protein (1908 nt) |
| MGG_00033 | -1.51815913 | Down | conserved hypothetical protein (1605 nt) |
| MGG_06347 | -1.514836414 | Down | conserved hypothetical protein (441 nt) |
| MGG_04922 | -1.512820264 | Down | alpha-N-arabinofuranosidase 2 (1083 nt) |
| MGG_01069 | -1.510674464 | Down | hypothetical protein (312 nt) |
| MGG_05692 | -1.509837762 | Down | conserved hypothetical protein (681 nt) |
| MGG_14793 | -1.507927117 | Down | hypothetical protein (306 nt) |
| MGG_12612 | -1.502500341 | Down | drug resistance transporter (1887 nt) |
| MGG_11560 | -1.501217831 | Down | taurine catabolism dioxygenase TauD (1125 nt) |
| MGG_09511 | -1.49128203 | Down | 2-nitropropane dioxygenase (990 nt) |
| MGG_07270 | -1.48244442 | Down | fatty aldehyde dehydrogenase (1647 nt) |
| MGG_14681 | -1.477507164 | Down | conserved hypothetical protein (1110 nt) |
| MGG_15174 | -1.476305437 | Down | predicted protein (342 nt) |
| MGG_14951 | -1.471998252 | Down | conserved hypothetical protein (3174 nt) |
| MGG_04248 | -1.471121655 | Down | conserved hypothetical protein (2223 nt) |
| MGG_05140 | -1.465508537 | Down | conserved hypothetical protein (1086 nt) |
| MGG_03245 | -1.461904104 | Down | aldose 1-epimerase (1215 nt) |
| MGG_00815 | -1.451773288 | Down | predicted protein (738 nt) |
| MGG_01993 | -1.450147629 | Down | predicted protein (306 nt) |
| MGG_10315 | -1.443282704 | Down | hydrophobin (339 nt) |
| MGG_13651 | -1.435823052 | Down | high-affinity glucose transporter (1605 nt) |
| MGG_09091 | -1.434294618 | Down | adiponectin receptor protein 1 (903 nt) |
| MGG_01456 | -1.433441793 | Down | conserved hypothetical protein (1674 nt) |
| MGG_10269 | -1.424797693 | Down | auxin Efflux Carrier superfamily (1818 nt) |
| MGG_04594 | -1.422470257 | Down | conserved hypothetical protein (1410 nt) |
| MGG_12899 | -1.422161934 | Down | DNA ligase 4 (3006 nt) |
| MGG_06260 | -1.418952549 | Down | conserved hypothetical protein (2727 nt) |
| MGG_03491 | -1.418713157 | Down | conserved hypothetical protein (1458 nt) |
| MGG_07211 | -1.418645279 | Down | conserved hypothetical protein (1482 nt) |
| MGG_08697 | -1.414463064 | Down | conserved hypothetical protein (3480 nt) |
| MGG_09533 | -1.412206292 | Down | predicted protein (501 nt) |
| MGG_09551 | -1.403741594 | Down | chitin synthase 3 (2673 nt) |
| MGG_12804 | -1.398733231 | Down | conserved hypothetical protein (2241 nt) |
| MGG_05871 | -1.397592365 | Down | integral membrane protein (1896 nt) |
| MGG_00268 | -1.396421821 | Down | conserved hypothetical protein (2049 nt) |
| MGG_09447 | -1.392923114 | Down | predicted protein (684 nt) |
| MGG_06364 | -1.390057478 | Down | hypothetical protein (1398 nt) |
| MGG_10452 | -1.388423507 | Down | predicted protein (756 nt) |
| MGG_09817 | -1.388052876 | Down | minor extracellular protease vpr (2664 nt) |
| MGG_05051 | -1.385121921 | Down | predicted protein (864 nt) |
| MGG_05114 | -1.380370367 | Down | hypothetical protein (1662 nt) |
| MGG_06863 | -1.379399163 | Down | conserved hypothetical protein (1134 nt) |
| MGG_09435 | -1.37926127 | Down | conserved hypothetical protein (1719 nt) |
| MGG_05722 | -1.378511623 | Down | conserved hypothetical protein (1755 nt) |
| MGG_02401 | -1.374703835 | Down | conserved hypothetical protein (1950 nt) |
| MGG_01673 | -1.373458396 | Down | conserved hypothetical protein (1788 nt) |
| MGG_00899 | -1.372926068 | Down | tubulin-specific chaperone C (1056 nt) |
| MGG_13008 | -1.370486472 | Down | predicted protein (384 nt) |
| MGG_04835 | -1.367605669 | Down | conserved hypothetical protein (1008 nt) |
| MGG_15050 | -1.367277082 | Down | conserved hypothetical protein (1203 nt) |
| MGG_11945 | -1.360317981 | Down | candidapepsin precursor (1560 nt) |
| MGG_06866 | -1.3594841 | Down | conserved hypothetical protein (759 nt) |
| MGG_06781 | -1.357079427 | Down | conserved hypothetical protein (1365 nt) |
| MGG_03392 | -1.357010554 | Down | cytochrome P450 11B1, mitochondrial precursor (1527 nt) |
| MGG_03941 | -1.350682695 | Down | conserved hypothetical protein (897 nt) |
| MGG_09444 | -1.34612779 | Down | methylitaconate delta2-delta3-isomerase (1278 nt) |
| MGG_10727 | -1.342332724 | Down | conserved hypothetical protein (1569 nt) |
| MGG_10585 | -1.340360376 | Down | monooxygenase (1008 nt) |
| MGG_10401 | -1.335858602 | Down | predicted protein (882 nt) |
| MGG_14590 | -1.327050328 | Down | conserved hypothetical protein (1344 nt) |
| MGG_09875 | -1.326221123 | Down | CAS1 domain-containing protein 1 (828 nt) |
| MGG_15134 | -1.324180547 | Down | conserved hypothetical protein (1029 nt) |
| MGG_01612 | -1.320419788 | Down | hypothetical protein (1818 nt) |
| MGG_05938 | -1.319541502 | Down | 2,2-dialkylglycine decarboxylase (1383 nt) |
| MGG_06888 | -1.315767594 | Down | glutamine synthetase (978 nt) |
| MGG_02081 | -1.3152412 | Down | conserved hypothetical protein (1638 nt) |
| MGG_01025 | -1.314815369 | Down | predicted protein (717 nt) |
| MGG_01311 | -1.313320922 | Down | nuclear elongation and deformation protein 1 (2298 nt) |
| MGG_12302 | -1.313253761 | Down | predicted protein (414 nt) |
| MGG_14205 | -1.306570073 | Down | conserved hypothetical protein (1107 nt) |
| MGG_08776 | -1.306195169 | Down | major myo-inositol transporter iolT (1899 nt) |
| MGG_04952 | -1.303477942 | Down | predicted protein (522 nt) |
| MGG_11239 | -1.303475332 | Down | RING finger protein (2259 nt) |
| MGG_03550 | -1.29736681 | Down | CDP-diacylglycerol-serine O-phosphatidyltransferase (729 nt) |
| MGG_15162 | -1.295210757 | Down | conserved hypothetical protein (1119 nt) |
| MGG_15012 | -1.283048195 | Down | conserved hypothetical protein (645 nt) |
| MGG_09271 | -1.282656701 | Down | conserved hypothetical protein (1092 nt) |
| MGG_07521 | -1.281401333 | Down | conserved hypothetical protein (1578 nt) |
| MGG_02273 | -1.280405227 | Down | hypothetical protein (576 nt) |
| MGG_13552 | -1.271732585 | Down | conserved hypothetical protein (1617 nt) |
| MGG_07887 | -1.269888077 | Down | 4-hydroxyacetophenone monooxygenase (1701 nt) |
| MGG_14925 | -1.267759394 | Down | hypothetical protein (180 nt) |
| MGG_00520 | -1.267642607 | Down | conserved hypothetical protein (357 nt) |
| MGG_13624 | -1.266786541 | Down | ABC transporter CDR4 (4644 nt) |
| MGG_08731 | -1.258589746 | Down | conserved hypothetical protein (2601 nt) |
| MGG_02583 | -1.256908303 | Down | mitochondrial ATP-binding cassette sub-family B member 6 (2385 nt) |
| MGG_07455 | -1.252739522 | Down | sorbitol utilization protein SOU2 (879 nt) |
| MGG_00819 | -1.252476214 | Down | niemann-Pick C1 protein precursor (3588 nt) |
| MGG_00426 | -1.246736876 | Down | short-chain dehydrogenase/reductase SDR (819 nt) |
| MGG_00747 | -1.246377062 | Down | palmitoyltransferase PFA4 (1488 nt) |
| MGG_05800 | -1.242628195 | Down | predicted protein (1188 nt) |
| MGG_00745 | -1.241375525 | Down | zn-dependent hydrolases (1233 nt) |
| MGG_00016 | -1.24082967 | Down | 2-epi-5-epi-valiolone synthase (1461 nt) |
| MGG_01094 | -1.238748592 | Down | conserved hypothetical protein (3573 nt) |
| MGG_10059 | -1.236333192 | Down | Gelsolin repeat-containing protein (1281 nt) |
| MGG_00623 | -1.233852328 | Down | hexokinase-1 (1686 nt) |
| MGG_02439 | -1.231528369 | Down | 2-nitropropane dioxygenase (1074 nt) |
| MGG_03029 | -1.229698609 | Down | 24 kDa metalloproteinase precursor (1074 nt) |
| MGG_01822 | -1.228057747 | Down | mitogen-activated protein kinase HOG1 (1074 nt) |
| MGG_00528 | -1.225885735 | Down | sterol esterase 2 (2268 nt) |
| MGG_04621 | -1.224401746 | Down | conserved hypothetical protein (1791 nt) |
| MGG_09562 | -1.222058619 | Down | conserved hypothetical protein (1608 nt) |
| MGG_01195 | -1.221596452 | Down | acetylesterase (732 nt) |
| MGG_10106 | -1.217591435 | Down | conserved hypothetical protein (1002 nt) |
| MGG_11308 | -1.209658931 | Down | zinc-binding alcohol dehydrogenase domain-containing protein 1 (1047 nt) |
| MGG_10400 | -1.209426856 | Down | glucan 1,3-beta-glucosidase (972 nt) |
| MGG_09639 | -1.206598264 | Down | cell wall alpha-1,3-glucan synthase ags1 (6756 nt) |
| MGG_01139 | -1.203413624 | Down | dihydroxy-acid dehydratase (1797 nt) |
| MGG_07482 | -1.200572679 | Down | conserved hypothetical protein (924 nt) |
| MGG_07856 | -1.20029865 | Down | pisatin demethylase (1542 nt) |
| MGG_04126 | -1.197056128 | Down | sulfate permease 2 (2370 nt) |
| MGG_10058 | -1.195351422 | Down | predicted protein (813 nt) |
| MGG_05976 | -1.190497206 | Down | transesterase (1218 nt) |
| MGG_03475 | -1.189435005 | Down | predicted protein (339 nt) |
| MGG_09802 | -1.185601801 | Down | conserved hypothetical protein (1734 nt) |
| MGG_13261 | -1.181160664 | Down | conserved hypothetical protein (738 nt) |
| MGG_14157 | -1.17995933 | Down | conserved hypothetical protein (1362 nt) |
| MGG_03070 | -1.174154936 | Down | epoxide hydrolase domain-containing protein (1233 nt) |
| MGG_08521 | -1.172603107 | Down | conserved hypothetical protein (1242 nt) |
| MGG_10525 | -1.170024378 | Down | conserved hypothetical protein (2520 nt) |
| MGG_01467 | -1.169925001 | Down | transmembrane protein 34 (1689 nt) |
| MGG_09238 | -1.168480139 | Down | conserved hypothetical protein (879 nt) |
| MGG_04177 | -1.166309419 | Down | conserved hypothetical protein (5589 nt) |
| MGG_04345 | -1.16083999 | Down | cytochrome P450 17A1 (1368 nt) |
| MGG_10083 | -1.14543044 | Down | endoglucanase 3 (1131 nt) |
| MGG_09371 | -1.145365428 | Down | phosphatidate cytidylyltransferase (1143 nt) |
| MGG_06593 | -1.143703628 | Down | endo-1,4-beta-xylanase 2 (771 nt) |
| MGG_05521 | -1.142717043 | Down | conserved hypothetical protein (1707 nt) |
| MGG_15245 | -1.137629446 | Down | conserved hypothetical protein (1161 nt) |
| MGG_14746 | -1.136383802 | Down | PH domain-containing protein (3543 nt) |
| MGG_11210 | -1.134496375 | Down | beta-glucosidase 1 (2631 nt) |
| MGG_06383 | -1.133974216 | Down | conserved hypothetical protein (717 nt) |
| MGG_02928 | -1.131083275 | Down | conserved hypothetical protein (1902 nt) |
| MGG_01883 | -1.129721257 | Down | conserved hypothetical protein (1632 nt) |
| MGG_09216 | -1.129532813 | Down | conserved hypothetical protein (456 nt) |
| MGG_01393 | -1.126896424 | Down | hypothetical protein (2214 nt) |
| MGG_08257 | -1.126617775 | Down | fatty acid transporter protein (1908 nt) |
| MGG_00190 | -1.125277221 | Down | conserved hypothetical protein (1263 nt) |
| MGG_06755 | -1.124478665 | Down | conserved hypothetical protein (1419 nt) |
| MGG_00829 | -1.122877586 | Down | alpha/beta hydrolase (972 nt) |
| MGG_08983 | -1.121015401 | Down | catechol 1,2-dioxygenase 1 (960 nt) |
| MGG_00562 | -1.120456325 | Down | NADPH-dependent 1-acyldihydroxyacetone phosphate reductase (855 nt) |
| MGG_13946 | -1.12042304 | Down | conserved hypothetical protein (1026 nt) |
| MGG_13334 | -1.118919584 | Down | general amino acid permease AGP2 (1731 nt) |
| MGG_08555 | -1.118181426 | Down | predicted protein (1470 nt) |
| MGG_10856 | -1.11064934 | Down | hypothetical protein (222 nt) |
| MGG_00939 | -1.110191576 | Down | conserved hypothetical protein (1425 nt) |
| MGG_15327 | -1.102653177 | Down | predicted protein (1059 nt) |
| MGG_07458 | -1.10240544 | Down | conserved hypothetical protein (2628 nt) |
| MGG_05499 | -1.101830675 | Down | predicted protein (822 nt) |
| MGG_11422 | -1.096263655 | Down | predicted protein (501 nt) |
| MGG_07369 | -1.095194126 | Down | conserved hypothetical protein (1269 nt) |
| MGG_07476 | -1.095180364 | Down | conserved hypothetical protein (2565 nt) |
| MGG_14632 | -1.094090285 | Down | hypothetical protein (216 nt) |
| MGG_02804 | -1.092397138 | Down | DNA repair protein rad16 (2883 nt) |
| MGG_09955 | -1.089511202 | Down | hypothetical protein (345 nt) |
| MGG_12317 | -1.089306541 | Down | hypothetical protein (864 nt) |
| MGG_01427 | -1.088881281 | Down | SET1 complex component ash2 (1704 nt) |
| MGG_07993 | -1.08604406 | Down | predicted protein (417 nt) |
| MGG_14639 | -1.081068424 | Down | predicted protein (1437 nt) |
| MGG_11597 | -1.079777841 | Down | glucosamine-fructose-6-phosphate aminotransferase (2121 nt) |
| MGG_02259 | -1.077846845 | Down | predicted protein (1590 nt) |
| MGG_12466 | -1.074085046 | Down | predicted protein (174 nt) |
| MGG_02444 | -1.07290822 | Down | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase 1 (3051 nt) |
| MGG_09478 | -1.072623134 | Down | hypothetical protein (561 nt) |
| MGG_13805 | -1.072440564 | Down | predicted protein (1032 nt) |
| MGG_07257 | -1.072244257 | Down | hypothetical protein (1407 nt) |
| MGG_08453 | -1.071790683 | Down | conserved hypothetical protein (1473 nt) |
| MGG_04002 | -1.069186812 | Down | hypothetical protein (204 nt) |
| MGG_08856 | -1.069185403 | Down | membrane-bound O-acyltransferase domain-containing protein 1 (1662 nt) |
| MGG_03351 | -1.065406996 | Down | glycosyl hydrolase (2391 nt) |
| MGG_10537 | -1.064899775 | Down | conserved hypothetical protein (1269 nt) |
| MGG_08774 | -1.064388508 | Down | chitin deacetylase (1107 nt) |
| MGG_01208 | -1.062644511 | Down | conserved hypothetical protein (639 nt) |
| MGG_12329 | -1.061240946 | Down | DNA excision repair protein ERCC-8 (1533 nt) |
| MGG_01653 | -1.056904511 | Down | nuclear transcription factor Y subunit B-3 (615 nt) |
| MGG_13944 | -1.054479897 | Down | hypothetical protein (288 nt) |
| MGG_06026 | -1.054080814 | Down | conserved hypothetical protein (1905 nt) |
| MGG_07734 | -1.053901938 | Down | M-phase inducer phosphatase (1710 nt) |
| MGG_01528 | -1.052273413 | Down | serine/threonine-protein phosphatase PP2A catalytic subunit (1320 nt) |
| MGG_02574 | -1.050021593 | Down | conserved hypothetical protein (2709 nt) |
| MGG_09949 | -1.046379935 | Down | oxysterol-binding protein (1572 nt) |
| MGG_13157 | -1.044105332 | Down | acetate kinase (1299 nt) |
| MGG_03649 | -1.043931126 | Down | protein phosphatase 1 regulatory subunit SDS22 (1146 nt) |
| MGG_09199 | -1.042885813 | Down | repressible acid phosphatase (1374 nt) |
| MGG_02423 | -1.03922131 | Down | ER lumen protein retaining receptor 2 (645 nt) |
| MGG_03219 | -1.038280996 | Down | conidiophore development protein hymA (963 nt) |
| MGG_04682 | -1.037803847 | Down | predicted protein (1278 nt) |
| MGG_00203 | -1.037786759 | Down | conserved hypothetical protein (1881 nt) |
| MGG_05714 | -1.037259619 | Down | conserved hypothetical protein (3984 nt) |
| MGG_01390 | -1.031395196 | Down | MFS hexose transporter (1533 nt) |
| MGG_00593 | -1.029202535 | Down | liver carboxylesterase (2088 nt) |
| MGG_06926 | -1.029146346 | Down | cyclopropane-fatty-acyl-phospholipid synthase (1056 nt) |
| MGG_04159 | -1.027834208 | Down | vacuolar calcium ion transporter (1749 nt) |
| MGG_01872 | -1.024833422 | Down | hypothetical protein (513 nt) |
| MGG_06331 | -1.022966687 | Down | conserved hypothetical protein (963 nt) |
| MGG_07569 | -1.022779869 | Down | cellobiose dehydrogenase (1728 nt) |
| MGG_10423 | -1.022063736 | Down | cellulase family protein (1269 nt) |
| MGG_08662 | -1.018657186 | Down | predicted protein (372 nt) |
| MGG_08201 | -1.018223292 | Down | DUF636 domain-containing protein (411 nt) |
| MGG_07872 | -1.017921908 | Down | conserved hypothetical protein (477 nt) |
| MGG_10492 | -1.017731319 | Down | mitochondrial carnitine carrier (1038 nt) |
| MGG_08367 | -1.017292236 | Down | conserved hypothetical protein (1071 nt) |
| MGG_00588 | -1.017142968 | Down | CIA30 family protein (702 nt) |
| MGG_12671 | -1.016753905 | Down | DUF718 domain-containing protein (447 nt) |
| MGG_13350 | -1.016571188 | Down | conserved hypothetical protein (1236 nt) |
| MGG_10482 | -1.015788886 | Down | conserved hypothetical protein (1218 nt) |
| MGG_03990 | -1.013649062 | Down | alpha-1,2 glucosyltransferase ALG10 (1983 nt) |
| MGG_02982 | -1.011866587 | Down | averantin oxidoreductase (1569 nt) |
| MGG_02542 | -1.008331275 | Down | predicted protein (2499 nt) |
| MGG_05527 | -1.006418938 | Down | peptidyl-tRNA hydrolase (648 nt) |
| MGG_08550 | -1.005932956 | Down | lipolytic enzyme (1083 nt) |
| MGG_00631 | -1.005719087 | Down | conserved hypothetical protein (1851 nt) |
| MGG_01550 | -1.005566676 | Down | neuronal calcium sensor 1 (624 nt) |
| MGG_09816 | -1.004576686 | Down | conserved hypothetical protein (933 nt) |
| MGG_07789 | -1.000708419 | Down | 3-phytase (2256 nt) |
新窗口打开
为分析DEGs与哪些生物学功能显著相关,利用GO注释及GO功能富集分析,对差异表达基因进行功能分类(图2-C)。结果显示,在细胞组分(cellular component)、分子功能(molecular function)和生物过程(biological process)3类功能注释的DEGs分别有118、299和308个。此外,细胞组分、分子功能和生物过程分别包含8、11和16个功能分类(图2-C)。其中,在细胞组分分组中,DEGs在细胞(cell; GO:0005623)和细胞区域(cell part;GO:0044464)类所占比例最高,大分子复合物(macromolecular complex;GO:0032991)类所占比例最低;在分子功能分组中,催化活性(catalytic activity;GO:0003824)和结合(binding;GO:0005488)类,DEGs所占比例最高,在结构分子活性(structural molecule activity;GO:0005198)和营养库活性类(nutrient reservoir activity;GO:0045735)所占比例最低;在生物过程分组中,DEGs在代谢过程(metabolic process;GO:0008152)和细胞过程(cellular process;GO:0009987)类所占比例最高,而在生物过程负调控(negative regulation of biological process;GO:0048519)类所占比例最低。
通过Kyoto Encyclopedia of Genes and Genomes(KEGG)pathway富集分析确定了DEGs参与的最主要生化代谢途径和信号转导途径[23],发现共有143个DEGs具有KEGG注释。结合注释结果,进行pathway富集分析,发现这些DEGs位于65个pathway中(附表2)。其中,DEGs位于代谢途径(metabolic pathways;pathway ID:ko04146)的数目最多(41个),其次位于次生代谢物质生物合成(biosynthesis of secondary metabolites;pathway ID:ko01110)(24个),甘油磷脂代谢(glycerophospholipid metabolism;pathway ID: ko00564)(6个),酵母MAPK信号通路(MAPK signaling pathway-yeast;pathway ID:ko04011)(5个),内质网对蛋白质的加工(protein processing in endoplasmic reticulum;pathway ID:ko04141)(5个),酵母细胞周期(cell cycle-yeast;pathway ID:ko04111)(5个)等途径中(附表2)。
Table S2
附表2
附表2143个差异表达基因的代谢途径
Table S2Pathways of the 143 differentially expressed genes
| 通路编号 Pathway ID | 描述 Description | Pathway注释的差异基因数 Number of DEGs with pathway annotation (143) | P-value |
|---|---|---|---|
| ko00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 3 (2.1%) | 0.01565294 |
| ko01110 | Biosynthesis of secondary metabolites | 24 (16.78%) | 0.03312813 |
| ko00564 | Glycerophospholipid metabolism | 6 (4.2%) | 0.04582877 |
| ko00051 | Fructose and mannose metabolism | 5 (3.5%) | 0.05442695 |
| ko00360 | Phenylalanine metabolism | 4 (2.8%) | 0.0633638 |
| ko00100 | Steroid biosynthesis | 4 (2.8%) | 0.07559576 |
| ko00650 | Butanoate metabolism | 5 (3.5%) | 0.07874347 |
| ko04650 | Natural killer cell mediated cytotoxicity | 1 (0.7%) | 0.09034359 |
| ko00311 | Penicillin and cephalosporin biosynthesis | 1 (0.7%) | 0.09034359 |
| ko00591 | Linoleic acid metabolism | 3 (2.1%) | 0.106024 |
| ko00565 | Ether lipid metabolism | 2 (1.4%) | 0.1186817 |
| ko03420 | Nucleotide excision repair | 4 (2.8%) | 0.1267428 |
| ko00640 | Propanoate metabolism | 3 (2.1%) | 0.1478173 |
| ko00500 | Starch and sucrose metabolism | 5 (3.5%) | 0.1640955 |
| ko04011 | MAPK signaling pathway - yeast | 5 (3.5%) | 0.1640955 |
| ko00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 2 (1.4%) | 0.1671517 |
| ko00910 | Nitrogen metabolism | 3 (2.1%) | 0.1704372 |
| ko00071 | Fatty acid metabolism | 3 (2.1%) | 0.2306433 |
| ko00300 | Lysine biosynthesis | 2 (1.4%) | 0.2705874 |
| ko00780 | Biotin metabolism | 1 (0.7%) | 0.2822858 |
| ko00010 | Glycolysis / Gluconeogenesis | 3 (2.1%) | 0.2940734 |
| ko04141 | Protein processing in endoplasmic reticulum | 5 (3.5%) | 0.3101778 |
| ko01100 | Metabolic pathways | 41 (28.67%) | 0.3105893 |
| ko04146 | Peroxisome | 4 (2.8%) | 0.3110427 |
| ko04144 | Endocytosis | 3 (2.1%) | 0.3198399 |
| ko03410 | Base excision repair | 2 (1.4%) | 0.3400542 |
| ko00230 | Purine metabolism | 6 (4.2%) | 0.3404561 |
| ko04113 | Meiosis - yeast | 5 (3.5%) | 0.3848884 |
| ko00250 | Alanine, aspartate and glutamate metabolism | 2 (1.4%) | 0.3908593 |
| ko00380 | Tryptophan metabolism | 3 (2.1%) | 0.3968992 |
| ko00062 | Fatty acid elongation | 1 (0.7%) | 0.4064083 |
| ko00290 | Valine, leucine and isoleucine biosynthesis | 1 (0.7%) | 0.4064083 |
| ko00310 | Lysine degradation | 2 (1.4%) | 0.4557394 |
| ko04111 | Cell cycle - yeast | 5 (3.5%) | 0.4957104 |
| ko04070 | Phosphatidylinositol signaling system | 1 (0.7%) | 0.5091888 |
| ko00061 | Fatty acid biosynthesis | 1 (0.7%) | 0.5319913 |
| ko00562 | Inositol phosphate metabolism | 1 (0.7%) | 0.5319913 |
| ko00240 | Pyrimidine metabolism | 3 (2.1%) | 0.5418366 |
| ko00561 | Glycerolipid metabolism | 2 (1.4%) | 0.5452004 |
| ko02010 | ABC transporters | 1 (0.7%) | 0.6131495 |
| ko03020 | RNA polymerase | 1 (0.7%) | 0.6131495 |
| ko00860 | Porphyrin and chlorophyll metabolism | 1 (0.7%) | 0.6311514 |
| ko00770 | Pantothenate and CoA biosynthesis | 1 (0.7%) | 0.6311514 |
| ko03430 | Mismatch repair | 1 (0.7%) | 0.6483211 |
| ko00410 | beta-Alanine metabolism | 1 (0.7%) | 0.6483211 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | 2 (1.4%) | 0.6592972 |
| ko00790 | Folate biosynthesis | 1 (0.7%) | 0.6803152 |
| ko01040 | Biosynthesis of unsaturated fatty acids | 1 (0.7%) | 0.6803152 |
| ko00480 | Glutathione metabolism | 1 (0.7%) | 0.6803152 |
| ko00510 | N-Glycan biosynthesis | 1 (0.7%) | 0.7094172 |
| ko00270 | Cysteine and methionine metabolism | 1 (0.7%) | 0.7094172 |
| ko00620 | Pyruvate metabolism | 1 (0.7%) | 0.7358868 |
| ko03022 | Basal transcription factors | 1 (0.7%) | 0.7482092 |
| ko00052 | Galactose metabolism | 1 (0.7%) | 0.7482092 |
| ko00460 | Cyanoamino acid metabolism | 1 (0.7%) | 0.7482092 |
| ko00280 | Valine, leucine and isoleucine degradation | 1 (0.7%) | 0.7599605 |
| ko00260 | Glycine, serine and threonine metabolism | 1 (0.7%) | 0.7711671 |
| ko03030 | DNA replication | 1 (0.7%) | 0.792045 |
| ko00340 | Histidine metabolism | 1 (0.7%) | 0.8017632 |
| ko00330 | Arginine and proline metabolism | 1 (0.7%) | 0.8830261 |
| ko03015 | mRNA surveillance pathway | 1 (0.7%) | 0.8937462 |
| ko03018 | RNA degradation | 1 (0.7%) | 0.9034902 |
| ko03010 | Ribosome | 2 (1.4%) | 0.9042958 |
| ko00350 | Tyrosine metabolism | 1 (0.7%) | 0.9508671 |
| ko03013 | RNA transport | 1 (0.7%) | 0.988561 |
新窗口打开
2.3 稻瘟病菌部分已知致病相关基因受Znf1调控
对Δznf1 RNA-Seq数据分析发现大约有20个已知的参与稻瘟病菌形态分化与致病性的基因受Znf1调控(表3)。其中,转录因子编码基因HOX7、FLBC以及脂质磷酸磷酸酶基因LPP2和LPP3在Δznf1中显著下调表达。此外,参与细胞渗透调节的MAPK(mitogen-activated protein kinase)基因OSM1和MAPKK基因PBS2,以及细胞壁形成相关基因CHS3和AGS1的表达也明显下调。然而,与一氧化氮合成和致病相关基因NOL3,参与黑色素合成相关基因ALB1、BUF1和RSY1以及编码热激蛋白40的MHF21的表达水平明显上调,说明在营养生长阶段Znf1对这些基因的转录可能起负调控作用。数据结果表明,这些与稻瘟病菌侵染相关形态分化、致病过程以及代谢相关基因可能直接或间接受Znf1调控。Table 3
表3
表3稻瘟病菌部分致病相关基因在Δznf1表达谱数据中的分析
Table 3Analysis of Δznf1 RNA-Seq data for several pathogenicity-related genes in M. oryzae
| 基因编号 Gene ID | 描述 Description | Log2 ratio (Δznf1/Guy11) | 参考文献 Reference |
|---|---|---|---|
| MGG_09994 | 脂质磷酸磷酸酶1 Lipid phosphate phosphatase 1 (Lpp3) | -3.2 | [24] |
| MGG_04699 | C2H2锌指结构蛋白FlbC C2H2 finger domain-containing protein FlbC (FlbC) | -2.4 | [25] |
| MGG_12865 | 保守假定蛋白 Conserved hypothetical protein (Hox7) | -2.4 | [16] |
| MGG_10268 | 丝裂原活化蛋白激酶激酶PBS2 MAP kinase kinase PBS2 (Pbs2) | -2.2 | [26] |
| MGG_02246 | 保守假定蛋白 Conserved hypothetical protein (Con6) | -1.7 | [27] |
| MGG_05988 | 脂质磷酸磷酸酶2 Lipid phosphate phosphatase 2 (Lpp2) | -1.5 | [24] |
| MGG_10315 | 疏水蛋白 Hydrophobin (Mpg1) | -1.4 | [28] |
| MGG_05871 | 膜整合蛋白 Integral membrane protein (Pth11) | -1.4 | [29] |
| MGG_09551 | 几丁质合成酶3 Chitin synthase 3 (Chs3) | -1.4 | [30] |
| MGG_01822 | 丝裂原活化蛋白激酶HOG1 MAP kinase HOG1 (Osm1) | -1.2 | [31] |
| MGG_09639 | 细胞壁α-1,3-葡聚糖合成酶Ags1 Cell wall alpha-1,3-glucan synthase Ags1 (Ags1) | -1.2 | [32] |
| MGG_10492 | 线粒体肉碱载体蛋白 Mitochondrial carnitine carrier (Crc1) | -1.0 | [33] |
| MGG_07953 | 双功能 P450:NADPH-P450 还原酶 Bifunctional P-450:NADPH-P450 reductase (Nol3) | 2.8 | [34] |
| MGG_07219 | 分生孢子黄色素合成聚酮合酶 Conidial yellow pigment biosynthesis polyketide synthase (Alb1) | 2.0 | [35-36] |
| MGG_02252 | 四羟基萘还原酶 Tetrahydroxynaphthalene reductase (Buf1) | 1.4 | [35-36] |
| MGG_08180 | 保守假定蛋白 Conserved hypothetical protein (Mhf21) | 1.7 | [37] |
| MGG_05059 | 小柱孢酮脱水酶 Scytalone dehydratase (Rsy1) | 1.2 | [35-36] |
| MGG_06131 | 保守假定蛋白 Conserved hypothetical protein (bZIP14) | 1.1 | [38] |
新窗口打开
2.4 受Znf1和Pmk1共同调控基因的分析
筛选Znf1和Pmk1共调控基因可为进一步有效鉴定Znf1下游调控基因以及明确Znf1与Pmk1的关系提供数据支持。因此,在同等条件下对Δpmk1进行了表达谱分析。将Δznf1与Δpmk1中的DEGs比较分析发现,Δznf1的410个下调表达基因中,有201个(约50%)基因在Δpmk1中的表达同样为下调;而Δznf1的299个上调基因中,有116个(约40%)基因在Δpmk1中也上调(图3,附表3)。比如,与在野生菌株Guy11中相比,LPP3以及HOX7都在Δznf1和Δpmk1中有相似的下调表达倍数,疏水蛋白基因MPG1在Δpmk1中显著下调表达(log2 ratio为-7.4),但在Δznf1 中MPG1仅下调表达2.5倍(log2 ratio为-1.4)。功能未知基因MGG_14997和MGG_03044在Δznf1和Δpmk1中均显著下调,log2 ratio(Δznf1/Guy11)和log2 ratio(Δpmk1/Guy11)分别为-12.1、-12.1、-4.3、-3.6,说明这两个基因受Znf1和Pmk1共同调控,且调控水平相似(附表3)。试验结果表明,Znf1可能作为Pmk1 MAPK信号途径下游的转录因子与Pmk1共同调控许多基因的转录过程。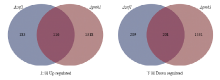 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图3Δznf1和Δpmk1中共同差异表达基因统计
-->Fig. 3Statistical analysis of differentially expressed genes in Δznf1 and Δpmk1
-->
Table S3
附表3
附表3Δznf1和Δpmk1菌丝中共同差异表达基因(|log2 ratio|≥1且 FDR<0.001)
Table S3Differentially expressed genes both in the mycelia of Δznf1和Δpmk1 (|log2 ratio|≥1且 FDR<0.001)
| 基因编号 Gene ID | Log2 ratio (Δpmk1/Guy11) | Log2 ratio (Δznf1/Guy11) | 上调-下调 Up-Down-Regulation | 描述 Description |
|---|---|---|---|---|
| MGG_15270 | 12.4252159 | 12.58871464 | Up | predicted protein (441 nt) |
| MGG_00585 | 11.09671515 | 11.93663794 | Up | predicted protein (354 nt) |
| MGG_05104 | 11.17990909 | 10.58871464 | Up | predicted protein (576 nt) |
| MGG_13037 | 8.906890596 | 9.550746785 | Up | conserved hypothetical protein (1692 nt) |
| MGG_12423 | 11.02236781 | 9.344295908 | Up | choline monooxygenase (1308 nt) |
| MGG_09972 | 10.79766153 | 9.276124405 | Up | conserved hypothetical protein (1605 nt) |
| MGG_00018 | 13.81478272 | 9.022367813 | Up | integral membrane protein (1650 nt) |
| MGG_08723 | 4.664689693 | 3.855717944 | Up | conserved hypothetical protein (972 nt) |
| MGG_14871 | 4.337869639 | 3.466699619 | Up | bifunctional molybdenum cofactor biosynthesis protein (801 nt) |
| MGG_04437 | 4.430042663 | 3.37914914 | Up | heat shock protein 78 (2421 nt) |
| MGG_12501 | 5.266786541 | 3.378511623 | Up | conserved hypothetical protein (1374 nt) |
| MGG_05084 | 3.820733952 | 3.328187085 | Up | conserved hypothetical protein (636 nt) |
| MGG_01989 | 4.266786541 | 3.266786541 | Up | predicted protein (1665 nt) |
| MGG_13116 | 1.015941544 | 3.062735755 | Up | conserved hypothetical protein (1260 nt) |
| MGG_09221 | 3.070389328 | 3.03562391 | Up | predicted protein (3054 nt) |
| MGG_06753 | 3.721099189 | 2.921840937 | Up | predicted protein (465 nt) |
| MGG_10110 | 2.338801913 | 2.884080713 | Up | conserved hypothetical protein (441 nt) |
| MGG_07952 | 1.819845866 | 2.865316383 | Up | predicted protein (516 nt) |
| MGG_10158 | 3.303780748 | 2.857980995 | Up | QDE-2-interacting protein (1713 nt) |
| MGG_05896 | 2.714653577 | 2.815494267 | Up | predicted protein (309 nt) |
| MGG_03158 | 3.718818247 | 2.604862058 | Up | pumilio-family RNA binding repeat protein (2328 nt) |
| MGG_05132 | 5.17194685 | 2.590701406 | Up | conserved hypothetical protein (1086 nt) |
| MGG_02059 | 1.432959407 | 2.523561956 | Up | conserved hypothetical protein (9060 nt) |
| MGG_03983 | 2.586844348 | 2.514368617 | Up | Hsp70 nucleotide exchange factor FES1 (654 nt) |
| MGG_02350 | 2.331205908 | 2.463656204 | Up | conserved hypothetical protein (927 nt) |
| MGG_10109 | 2.445516154 | 2.438920966 | Up | conserved hypothetical protein (465 nt) |
| MGG_09138 | 5.678738771 | 2.42053125 | Up | glutathione S-transferase II (672 nt) |
| MGG_07404 | 1.943416472 | 2.387023123 | Up | tripeptidyl-peptidase 1 (1806 nt) |
| MGG_09139 | 5.358628777 | 2.38669835 | Up | laccase-1 (1794 nt) |
| MGG_06759 | 2.044335357 | 2.364189581 | Up | heat shock protein 80 (2109 nt) |
| MGG_10271 | 2.478047297 | 2.342392197 | Up | conserved hypothetical protein (3960 nt) |
| MGG_02436 | 2.489221598 | 2.296338285 | Up | conserved hypothetical protein (459 nt) |
| MGG_10415 | 2.795180208 | 2.270089163 | Up | predicted protein (2403 nt) |
| MGG_06459 | 2.679268795 | 2.200152894 | Up | heat shock protein HSP98 (2781 nt) |
| MGG_07949 | 2.485303261 | 2.170357399 | Up | conserved hypothetical protein (879 nt) |
| MGG_10108 | 1.898958651 | 2.167556766 | Up | conserved hypothetical protein (789 nt) |
| MGG_03115 | 3.651338443 | 2.145452682 | Up | predicted protein (1401 nt) |
| MGG_06958 | 2.5667168 | 2.06641867 | Up | hsp70-like protein (1956 nt) |
| MGG_02372 | 1.805673786 | 2.018018704 | Up | hypothetical protein (729 nt) |
| MGG_08424 | 2.698303974 | 2.0138058 | Up | endo-1,4-beta-xylanase I (696 nt) |
| MGG_14351 | 2.667083195 | 1.94240208 | Up | glycosyltransferase (1269 nt) |
| MGG_14340 | 1.43300209 | 1.934604368 | Up | predicted protein (324 nt) |
| MGG_15049 | 2.29820072 | 1.871675903 | Up | predicted protein (1944 nt) |
| MGG_02267 | 4.455568979 | 1.866733469 | Up | predicted protein (1044 nt) |
| MGG_10683 | 3.971543554 | 1.849665727 | Up | predicted protein (1479 nt) |
| MGG_03238 | 2.754089772 | 1.848445436 | Up | zinc finger protein ZPR1 (1533 nt) |
| MGG_02095 | 3.391566506 | 1.839416707 | Up | conserved hypothetical protein (996 nt) |
| MGG_08378 | 2.876341532 | 1.824913293 | Up | cytochrome P450 oxidoreductase GliF (1605 nt) |
| MGG_04511 | 2.095611912 | 1.821029859 | Up | MFS transporter (1461 nt) |
| MGG_12697 | 2.509796533 | 1.798601538 | Up | pheromone-regulated membrane protein 10 (2973 nt) |
| MGG_05505 | 1.13606155 | 1.795986108 | Up | autophagy protein (Atg22) (1704 nt) |
| MGG_01410 | 4.081388327 | 1.791820579 | Up | glutathione S-transferase Gst3 (1035 nt) |
| MGG_02043 | 3.251588514 | 1.777192952 | Up | BTB/POZ domain-containing protein (510 nt) |
| MGG_14675 | 2.084092679 | 1.772956761 | Up | hypothetical protein (180 nt) |
| MGG_06616 | 1.146841388 | 1.762500686 | Up | high-affinity methionine permease (1833 nt) |
| MGG_02096 | 2.378511623 | 1.757618704 | Up | glycoside hydrolase (2478 nt) |
| MGG_02002 | 2.710465401 | 1.744835012 | Up | conserved hypothetical protein (1281 nt) |
| MGG_14621 | 5.47952576 | 1.724556807 | Up | conserved hypothetical protein (279 nt) |
| MGG_11336 | 1.68772515 | 1.703898264 | Up | conserved hypothetical protein (774 nt) |
| MGG_05515 | 4.516015147 | 1.669026766 | Up | predicted protein (588 nt) |
| MGG_08989 | 2.02379212 | 1.660472735 | Up | conserved hypothetical protein (816 nt) |
| MGG_13019 | 1.288469453 | 1.631716269 | Up | predicted protein (690 nt) |
| MGG_11680 | 3.174174474 | 1.620690205 | Up | AAA family ATPase (2112 nt) |
| MGG_00791 | 2.343677678 | 1.603109847 | Up | Lactamase_B domain containing protein (1077 nt) |
| MGG_04627 | 2.017921908 | 1.566815154 | Up | conserved hypothetical protein (2787 nt) |
| MGG_08519 | 4.717022911 | 1.565654069 | Up | aflatoxin B1 aldehyde reductase member 3 (1002 nt) |
| MGG_12714 | 3.215904085 | 1.563004767 | Up | conserved hypothetical protein (930 nt) |
| MGG_13562 | 2.936230497 | 1.541985428 | Up | conserved hypothetical protein (1593 nt) |
| MGG_01454 | 2.096343869 | 1.541813265 | Up | conserved hypothetical protein (720 nt) |
| MGG_01364 | 3.189777559 | 1.531668886 | Up | conserved hypothetical protein (2925 nt) |
| MGG_08980 | 1.781051303 | 1.528536579 | Up | stress-induced-phosphoprotein 1 (1755 nt) |
| MGG_11991 | 2.58090102 | 1.516744272 | Up | predicted protein (252 nt) |
| MGG_07766 | 1.874973469 | 1.514203204 | Up | predicted protein (372 nt) |
| MGG_04736 | 2.432603231 | 1.500318333 | Up | conserved hypothetical protein (912 nt) |
| MGG_05968 | 1.502985604 | 1.483038711 | Up | endoribonuclease L-PSP (411 nt) |
| MGG_02977 | 1.278859373 | 1.479542529 | Up | predicted protein (1809 nt) |
| MGG_02854 | 2.907591239 | 1.47566256 | Up | conserved hypothetical protein (942 nt) |
| MGG_08726 | 1.570262889 | 1.47279708 | Up | hypothetical protein (588 nt) |
| MGG_00809 | 1.45172962 | 1.472177608 | Up | predicted protein (1770 nt) |
| MGG_11779 | 1.443301351 | 1.447112965 | Up | conserved hypothetical protein (5598 nt) |
| MGG_07402 | 1.446512239 | 1.436863862 | Up | conserved hypothetical protein (465 nt) |
| MGG_09019 | 1.860437934 | 1.3852473 | Up | secretory phospholipase A2 (561 nt) |
| MGG_07078 | 2.571377145 | 1.347104217 | Up | DUF775 domain-containing protein (609 nt) |
| MGG_00156 | 4.616608127 | 1.330364932 | Up | conserved hypothetical protein (966 nt) |
| MGG_00963 | 2.119999776 | 1.327804661 | Up | predicted protein (2850 nt) |
| MGG_05574 | 2.463885759 | 1.292180751 | Up | conserved hypothetical protein (1539 nt) |
| MGG_08029 | 1.175849835 | 1.281729267 | Up | conserved hypothetical protein (942 nt) |
| MGG_11682 | 2.598873395 | 1.281189416 | Up | phenylacetone monooxygenase (1674 nt) |
| MGG_00282 | 2.187627003 | 1.258104518 | Up | conserved hypothetical protein (2610 nt) |
| MGG_08104 | 1.480588488 | 1.254630144 | Up | 41 kDa peptidyl-prolyl cis-trans isomerase (1131 nt) |
| MGG_01951 | 2.831080992 | 1.25334101 | Up | 4-coumarate-CoA ligase 2 (2010 nt) |
| MGG_08158 | 1.466117407 | 1.217117479 | Up | predicted protein (993 nt) |
| MGG_05978 | 1.070591968 | 1.216910356 | Up | conserved hypothetical protein (564 nt) |
| MGG_05913 | 2.441623161 | 1.213091603 | Up | hypothetical protein (657 nt) |
| MGG_04757 | 1.824771975 | 1.193739826 | Up | conserved hypothetical protein (1485 nt) |
| MGG_04346 | 1.993886716 | 1.189968987 | Up | sterol 24-C-methyltransferase (1173 nt) |
| MGG_05611 | 2.44908666 | 1.181656367 | Up | hypothetical protein (243 nt) |
| MGG_00834 | 1.225326394 | 1.136488996 | Up | hypothetical protein (987 nt) |
| MGG_14857 | 1.379032546 | 1.126377286 | Up | conserved hypothetical protein (1050 nt) |
| MGG_02105 | 2.84595717 | 1.112563575 | Up | conserved hypothetical protein (915 nt) |
| MGG_03005 | 1.209718591 | 1.109080361 | Up | conserved hypothetical protein (1332 nt) |
| MGG_08007 | 1.599353593 | 1.107108893 | Up | conserved hypothetical protein (762 nt) |
| MGG_14651 | 1.68921535 | 1.08767481 | Up | conserved hypothetical protein (696 nt) |
| MGG_04242 | 1.610501769 | 1.081893423 | Up | bax Inhibitor family protein (786 nt) |
| MGG_09664 | 1.329214449 | 1.075597619 | Up | beta-mannosidase (2739 nt) |
| MGG_06131 | 1.292159087 | 1.073159746 | Up | conserved hypothetical protein (720 nt) |
| MGG_05940 | 3.869107257 | 1.070943772 | Up | short chain dehydrogenase (780 nt) |
| MGG_10613 | 1.170277265 | 1.054447784 | Up | retinol dehydrogenase 11 (975 nt) |
| MGG_08142 | 3.902461751 | 1.052911531 | Up | conserved hypothetical protein (1305 nt) |
| MGG_03098 | 2.155595633 | 1.049687124 | Up | conserved hypothetical protein (984 nt) |
| MGG_09328 | 3.202101303 | 1.048429433 | Up | conserved hypothetical protein (1113 nt) |
| MGG_04569 | 3.895608462 | 1.029318937 | Up | NADPH dehydrogenase (1329 nt) |
| MGG_00738 | 1.481889031 | 1.025321666 | Up | zinc-binding alcohol dehydrogenase domain-containing protein 2 (1020 nt) |
| MGG_02355 | 1.947266884 | 1.018265026 | Up | predicted protein (687 nt) |
| MGG_02445 | 2.016601967 | 1.008878151 | Up | predicted protein (801 nt) |
| MGG_01692 | 1.496153234 | 1.008721271 | Up | conserved hypothetical protein (1206 nt) |
| MGG_14668 | -3.298015475 | -12.80976813 | Down | hypothetical protein (228 nt) |
| MGG_11005 | -2.198904068 | -12.36632221 | Down | conserved hypothetical protein (1368 nt) |
| MGG_14997 | -12.14210706 | -12.14210706 | Down | hypothetical protein (219 nt) |
| MGG_15409 | -1.630445271 | -4.635588574 | Down | predicted protein (432 nt) |
| MGG_05719 | -3.374964882 | -2.05779793 | Down | 30 kDa heat shock protein (723 nt) |
| MGG_03044 | -3.610359278 | -4.310142181 | Down | conserved hypothetical protein (1101 nt) |
| MGG_02942 | -1.468094002 | -4.161723188 | Down | conserved hypothetical protein (1749 nt) |
| MGG_07807 | -1.828888084 | -4.138216142 | Down | conserved hypothetical protein (648 nt) |
| MGG_09879 | -3.161857344 | -3.992938336 | Down | conserved hypothetical protein (438 nt) |
| MGG_00766 | -2.934617277 | -3.619372938 | Down | conserved hypothetical protein (558 nt) |
| MGG_07857 | -4.96133838 | -3.413850584 | Down | conserved hypothetical protein (1125 nt) |
| MGG_10650 | -5.107637884 | -3.408267701 | Down | hypothetical protein (291 nt) |
| MGG_03825 | -9.764871591 | -3.273018494 | Down | isotrichodermin C-15 hydroxylase (1617 nt) |
| MGG_05250 | -1.389007222 | -3.245205238 | Down | conserved hypothetical protein (2520 nt) |
| MGG_09994 | -2.903078982 | -3.174512271 | Down | lipid phosphate phosphatase 1 (1140 nt) |
| MGG_02290 | -3.545532356 | -3.141142101 | Down | conserved hypothetical protein (858 nt) |
| MGG_01391 | -5.276496666 | -3.070166018 | Down | ent-kaurene oxidase (1569 nt) |
| MGG_09418 | -1.14038108 | -3.02188266 | Down | conserved hypothetical protein (429 nt) |
| MGG_08971 | -3.422138556 | -2.886958693 | Down | conserved hypothetical protein (573 nt) |
| MGG_03329 | -2.624176135 | -3.169891506 | Down | small heat shock protein (633 nt) |
| MGG_08326 | -2.827301286 | -2.881995905 | Down | conserved hypothetical protein (1416 nt) |
| MGG_10322 | -1.926158373 | -2.87570589 | Down | cortical patch protein (765 nt) |
| MGG_09134 | -6.916311635 | -2.765762647 | Down | hypothetical protein (357 nt) |
| MGG_04171 | -2.600160554 | -2.721732314 | Down | predicted protein (1050 nt) |
| MGG_14581 | -2.126487637 | -2.718799594 | Down | hypothetical protein (744 nt) |
| MGG_07348 | -2.099094415 | -2.697353738 | Down | predicted protein (420 nt) |
| MGG_10713 | -4.412028749 | -2.660707862 | Down | conserved hypothetical protein (813 nt) |
| MGG_02329 | -2.776515741 | -2.649306721 | Down | isotrichodermin C-15 hydroxylase (1632 nt) |
| MGG_00733 | -2.378276173 | -2.605714662 | Down | conserved hypothetical protein (1026 nt) |
| MGG_12800 | -2.149777312 | -2.531615468 | Down | conserved hypothetical protein (1257 nt) |
| MGG_01152 | -2.06786603 | -2.496200352 | Down | conserved hypothetical protein (549 nt) |
| MGG_07871 | -3.074075099 | -2.493092449 | Down | conserved hypothetical protein (726 nt) |
| MGG_08498 | -4.181479228 | -2.492768803 | Down | isotrichodermin C-15 hydroxylase (1506 nt) |
| MGG_07697 | -2.947300243 | -2.470862199 | Down | superoxide dismutase (642 nt) |
| MGG_12865 | -1.990940661 | -2.412767326 | Down | conserved hypothetical protein (1344 nt) |
| MGG_11876 | -1.968229221 | -2.393623513 | Down | sugar transporter STL1 (1722 nt) |
| MGG_01368 | -2.284535475 | -2.345597001 | Down | conserved hypothetical protein (714 nt) |
| MGG_10714 | -5.989415578 | -2.315031003 | Down | cyanide hydratase (1071 nt) |
| MGG_03977 | -1.306035559 | -2.298702913 | Down | hypothetical protein (1389 nt) |
| MGG_01369 | -3.381956573 | -2.295885623 | Down | alpha/beta hydrolase fold-3 domain-containing protein (1044 nt) |
| MGG_10429 | -1.969063833 | -2.277318632 | Down | conserved hypothetical protein (963 nt) |
| MGG_05804 | -3.656239914 | -2.269857074 | Down | phospholipase D p1 (2574 nt) |
| MGG_13655 | -2.325562743 | -2.268229568 | Down | conserved hypothetical protein (1416 nt) |
| MGG_09445 | -1.745816512 | -2.266500219 | Down | conserved hypothetical protein (1437 nt) |
| MGG_07088 | -1.983069604 | -2.257691984 | Down | hypothetical protein (315 nt) |
| MGG_09473 | -2.923306997 | -2.24858331 | Down | hypothetical protein (354 nt) |
| MGG_09000 | -11.72280753 | -2.230954435 | Down | protein kinase domain-containing protein (2160 nt) |
| MGG_08405 | -4.502500341 | -2.190222416 | Down | retinol dehydrogenase 13 (969 nt) |
| MGG_06545 | -1.491190941 | -2.168411949 | Down | conserved hypothetical protein (2358 nt) |
| MGG_08439 | -2.822122796 | -2.161473337 | Down | predicted protein (846 nt) |
| MGG_13198 | -2.155937856 | -2.159984689 | Down | predicted protein (270 nt) |
| MGG_01542 | -1.740912779 | -2.072756342 | Down | endo-1,4-beta-xylanase (996 nt) |
| MGG_09851 | -1.605426603 | -2.05288558 | Down | predicted protein (1029 nt) |
| MGG_12366 | -1.629422558 | -2.051002903 | Down | hypothetical protein (162 nt) |
| MGG_07356 | -3.355480655 | -2.050626073 | Down | predicted protein (1701 nt) |
| MGG_10503 | -1.83865173 | -2.038908043 | Down | mannitol-1-phosphate 5-dehydrogenase (1203 nt) |
| MGG_21009 | -1.862947248 | -2.035128223 | Down | mitochondria cytochrome c oxidase subunit I cox1 (1599 nt) |
| MGG_02709 | -4.08516956 | -2.016440916 | Down | conserved hypothetical protein (465 nt) |
| MGG_15428 | -1.279201609 | -2.012748493 | Down | para-nitrobenzyl esterase (1674 nt) |
| MGG_03764 | -1.207232614 | -2 | Down | salicylate hydroxylase (1395 nt) |
| MGG_08059 | -3.573950442 | -1.99175344 | Down | conserved hypothetical protein (615 nt) |
| MGG_02803 | -1.994865686 | -1.972238488 | Down | hypothetical protein (207 nt) |
| MGG_04251 | -1.062442879 | -1.96800463 | Down | phosphate transporter family protein (1782 nt) |
| MGG_09001 | -5.224001674 | -1.915662644 | Down | hypothetical protein (474 nt) |
| MGG_00114 | -2.379300001 | -1.914793825 | Down | conserved hypothetical protein (594 nt) |
| MGG_00993 | -3.513254318 | -1.89630787 | Down | vacuolar iron transporter Ccc1 (1014 nt) |
| MGG_03643 | -1.63169993 | -1.885795281 | Down | conserved hypothetical protein (3816 nt) |
| MGG_01373 | -1.56802005 | -1.846069502 | Down | arabinose-proton symporter (1728 nt) |
| MGG_07806 | -5.134649527 | -1.800230488 | Down | conserved hypothetical protein (1230 nt) |
| MGG_04203 | -1.078328361 | -1.799556551 | Down | 4-coumarate-CoA ligase 1 (1671 nt) |
| MGG_01544 | -2.897819605 | -1.788997336 | Down | pisatin demethylase (1464 nt) |
| MGG_14146 | -13.98726401 | -1.764469109 | Down | hypothetical protein (384 nt) |
| MGG_09828 | -1.149050919 | -1.763810581 | Down | vacuolar conductance protein (1959 nt) |
| MGG_02752 | -2.678576833 | -1.734749061 | Down | hypothetical protein (1071 nt) |
| MGG_05358 | -1.222148207 | -1.714428705 | Down | LYR family protein (267 nt) |
| MGG_09323 | -2.623386485 | -1.703160095 | Down | phenylacetone monooxygenase (1944 nt) |
| MGG_09260 | -1.5966815 | -1.697890657 | Down | integral membrane protein (495 nt) |
| MGG_13798 | -14.07430874 | -1.686291456 | Down | predicted protein (915 nt) |
| MGG_07966 | -1.256055394 | -1.684741955 | Down | phosphate transporter family protein (1767 nt) |
| MGG_02929 | -1.30712216 | -1.675273272 | Down | phosphotransferase enzyme family protein (789 nt) |
| MGG_10359 | -2.127489735 | -1.673771768 | Down | conserved hypothetical protein (1248 nt) |
| MGG_04257 | -1.213955584 | -1.672160943 | Down | secretory lipase family protein (1269 nt) |
| MGG_07890 | -1.490184276 | -1.667424661 | Down | aldehyde dehydrogenase 3I1 (1482 nt) |
| MGG_02332 | -5.967706597 | -1.661898167 | Down | conserved hypothetical protein (810 nt) |
| MGG_01376 | -1.203750015 | -1.657020649 | Down | tyrosine-protein phosphatase 1 (3420 nt) |
| MGG_01945 | -1.348094132 | -1.653849412 | Down | predicted protein (729 nt) |
| MGG_10973 | -2.155358888 | -1.647520814 | Down | conserved hypothetical protein (570 nt) |
| MGG_10422 | -6.573787425 | -1.646850887 | Down | C6 transcription factor OefC (2112 nt) |
| MGG_09985 | -2.729103476 | -1.628174568 | Down | hypothetical protein (1347 nt) |
| MGG_14578 | -1.750480401 | -1.620712526 | Down | predicted protein (855 nt) |
| MGG_13428 | -1.222885988 | -1.605463615 | Down | mitochondrial phosphate carrier protein (945 nt) |
| MGG_01798 | -1.143330494 | -1.587742596 | Down | conserved hypothetical protein (906 nt) |
| MGG_12664 | -1.927149435 | -1.579226131 | Down | conserved hypothetical protein (777 nt) |
| MGG_06181 | -2.643544222 | -1.578735926 | Down | predicted protein (1539 nt) |
| MGG_01463 | -1.577173699 | -1.576467707 | Down | phosphate metabolism protein 7 (2682 nt) |
| MGG_10092 | -3.783980414 | -1.574527048 | Down | hypothetical protein (1086 nt) |
| MGG_00097 | -2.915493244 | -1.569986896 | Down | conserved hypothetical protein (972 nt) |
| MGG_10394 | -4.911891277 | -1.553362167 | Down | conserved hypothetical protein (618 nt) |
| MGG_10053 | -2.443347895 | -1.552972386 | Down | conserved hypothetical protein (867 nt) |
| MGG_15228 | -2.91083778 | -1.538419915 | Down | predicted protein (354 nt) |
| MGG_05897 | -1.372405891 | -1.535135392 | Down | predicted protein (462 nt) |
| MGG_00830 | -2.72106473 | -1.519972879 | Down | conserved hypothetical protein (1908 nt) |
| MGG_00033 | -2.15631457 | -1.51815913 | Down | conserved hypothetical protein (1605 nt) |
| MGG_04922 | -2.498173488 | -1.512820264 | Down | alpha-N-arabinofuranosidase 2 (1083 nt) |
| MGG_05692 | -2.055684133 | -1.509837762 | Down | conserved hypothetical protein (681 nt) |
| MGG_14681 | -2.293223008 | -1.477507164 | Down | conserved hypothetical protein (1110 nt) |
| MGG_15174 | -1.676243008 | -1.476305437 | Down | predicted protein (342 nt) |
| MGG_05140 | -1.039275149 | -1.465508537 | Down | conserved hypothetical protein (1086 nt) |
| MGG_00815 | -4.545589327 | -1.451773288 | Down | predicted protein (738 nt) |
| MGG_01993 | -4.432702494 | -1.450147629 | Down | predicted protein (306 nt) |
| MGG_10315 | -7.432355637 | -1.443282704 | Down | hydrophobin (339 nt) |
| MGG_04594 | -1.06210985 | -1.422470257 | Down | conserved hypothetical protein (1410 nt) |
| MGG_06260 | -3.62058641 | -1.418952549 | Down | conserved hypothetical protein (2727 nt) |
| MGG_07211 | -1.990331352 | -1.418645279 | Down | conserved hypothetical protein (1482 nt) |
| MGG_05871 | -2.888578717 | -1.397592365 | Down | integral membrane protein (1896 nt) |
| MGG_00268 | -1.301002256 | -1.396421821 | Down | conserved hypothetical protein (2049 nt) |
| MGG_09447 | -3.113863284 | -1.392923114 | Down | predicted protein (684 nt) |
| MGG_06364 c2h2 | -2.305329222 | -1.390057478 | Down | hypothetical protein (1398 nt) |
| MGG_10452 | -1.258362966 | -1.388423507 | Down | predicted protein (756 nt) |
| MGG_05051 | -1.568568062 | -1.385121921 | Down | predicted protein (864 nt) |
| MGG_05114 | -1.957295549 | -1.380370367 | Down | hypothetical protein (1662 nt) |
| MGG_06863 | -1.446733214 | -1.379399163 | Down | conserved hypothetical protein (1134 nt) |
| MGG_00899 | -1.038913905 | -1.372926068 | Down | tubulin-specific chaperone C (1056 nt) |
| MGG_13008 | -4.824004051 | -1.370486472 | Down | predicted protein (384 nt) |
| MGG_04835 | -1.101212251 | -1.367605669 | Down | conserved hypothetical protein (1008 nt) |
| MGG_11945 | -1.370706095 | -1.360317981 | Down | candidapepsin precursor (1560 nt) |
| MGG_06781 | -1.471499338 | -1.357079427 | Down | conserved hypothetical protein (1365 nt) |
| MGG_03392 | -3.572023445 | -1.357010554 | Down | cytochrome P450 11B1, mitochondrial precursor (1527 nt) |
| MGG_03941 | -2.031093294 | -1.350682695 | Down | conserved hypothetical protein (897 nt) |
| MGG_15134 | -1.513608912 | -1.324180547 | Down | conserved hypothetical protein (1029 nt) |
| MGG_01612 | -2.97012947 | -1.320419788 | Down | hypothetical protein (1818 nt) |
| MGG_06888 | -4.18999471 | -1.315767594 | Down | glutamine synthetase (978 nt) |
| MGG_02081 | -1.13884463 | -1.3152412 | Down | conserved hypothetical protein (1638 nt) |
| MGG_01025 | -1.566072607 | -1.314815369 | Down | predicted protein (717 nt) |
| MGG_12302 | -3.623436649 | -1.313253761 | Down | predicted protein (414 nt) |
| MGG_14205 | -1.897111208 | -1.306570073 | Down | conserved hypothetical protein (1107 nt) |
| MGG_07521 | -2.054563812 | -1.281401333 | Down | conserved hypothetical protein (1578 nt) |
| MGG_02273 | -8.206429711 | -1.280405227 | Down | hypothetical protein (576 nt) |
| MGG_13552 | -1.036846026 | -1.271732585 | Down | conserved hypothetical protein (1617 nt) |
| MGG_07887 | -2.492280498 | -1.269888077 | Down | 4-hydroxyacetophenone monooxygenase (1701 nt) |
| MGG_07455 | -1.525919085 | -1.252739522 | Down | sorbitol utilization protein SOU2 (879 nt) |
| MGG_00819 | -1.540981307 | -1.252476214 | Down | niemann-Pick C1 protein precursor (3588 nt) |
| MGG_00016 | -1.47720632 | -1.24082967 | Down | 2-epi-5-epi-valiolone synthase (1461 nt) |
| MGG_10059 | -2.754979047 | -1.236333192 | Down | Gelsolin repeat-containing protein (1281 nt) |
| MGG_00528 | -1.060745796 | -1.225885735 | Down | sterol esterase 2 (2268 nt) |
| MGG_01195 | -1.057252482 | -1.221596452 | Down | acetylesterase (732 nt) |
| MGG_01139 | -1.261454236 | -1.203413624 | Down | dihydroxy-acid dehydratase (1797 nt) |
| MGG_07482 | -1.788337696 | -1.200572679 | Down | conserved hypothetical protein (924 nt) |
| MGG_07856 | -1.736965594 | -1.20029865 | Down | pisatin demethylase (1542 nt) |
| MGG_04126 | -3.325886108 | -1.197056128 | Down | sulfate permease 2 (2370 nt) |
| MGG_10058 | -1.574549094 | -1.195351422 | Down | predicted protein (813 nt) |
| MGG_05976 | -1.212222528 | -1.190497206 | Down | transesterase (1218 nt) |
| MGG_03475 | -4.5325338 | -1.189435005 | Down | predicted protein (339 nt) |
| MGG_09802 | -2.41739292 | -1.185601801 | Down | conserved hypothetical protein (1734 nt) |
| MGG_03070 | -3.115573215 | -1.174154936 | Down | epoxide hydrolase domain-containing protein (1233 nt) |
| MGG_08521 | -1.865223321 | -1.172603107 | Down | conserved hypothetical protein (1242 nt) |
| MGG_10525 | -1.593134174 | -1.170024378 | Down | conserved hypothetical protein (2520 nt) |
| MGG_04345 | -3.438620981 | -1.16083999 | Down | cytochrome P450 17A1 (1368 nt) |
| MGG_10083 | -1.254364811 | -1.14543044 | Down | endoglucanase 3 (1131 nt) |
| MGG_09371 | -1.111678983 | -1.145365428 | Down | phosphatidate cytidylyltransferase (1143 nt) |
| MGG_06593 | -4.186067212 | -1.143703628 | Down | endo-1,4-beta-xylanase 2 (771 nt) |
| MGG_05521 | -1.41214922 | -1.142717043 | Down | conserved hypothetical protein (1707 nt) |
| MGG_11210 | -1.409390936 | -1.134496375 | Down | beta-glucosidase 1 (2631 nt) |
| MGG_01883 | -1.599360469 | -1.129721257 | Down | conserved hypothetical protein (1632 nt) |
| MGG_06755 | -1.610663011 | -1.124478665 | Down | conserved hypothetical protein (1419 nt) |
| MGG_00829 | -2.409350363 | -1.122877586 | Down | alpha/beta hydrolase (972 nt) |
| MGG_08983 | -2.207727034 | -1.121015401 | Down | catechol 1,2-dioxygenase 1 (960 nt) |
| MGG_13946 | -1.113996771 | -1.12042304 | Down | conserved hypothetical protein (1026 nt) |
| MGG_13334 | -2.139981199 | -1.118919584 | Down | general amino acid permease AGP2 (1731 nt) |
| MGG_00939 | -2.060144203 | -1.110191576 | Down | conserved hypothetical protein (1425 nt) |
| MGG_14632 | -5.437663374 | -1.094090285 | Down | hypothetical protein (216 nt) |
| MGG_02804 | -1.215728691 | -1.092397138 | Down | DNA repair protein rad16 (2883 nt) |
| MGG_09955 | -1.969723867 | -1.089511202 | Down | hypothetical protein (345 nt) |
| MGG_12317 | -12.93479659 | -1.089306541 | Down | hypothetical protein (864 nt) |
| MGG_07993 | -7.271589209 | -1.08604406 | Down | predicted protein (417 nt) |
| MGG_14639 | -1.974319842 | -1.081068424 | Down | predicted protein (1437 nt) |
| MGG_02259 | -1.403222136 | -1.077846845 | Down | predicted protein (1590 nt) |
| MGG_12466 | -1.067343468 | -1.074085046 | Down | predicted protein (174 nt) |
| MGG_08453 | -2.110264831 | -1.071790683 | Down | conserved hypothetical protein (1473 nt) |
| MGG_03351 | -1.431244789 | -1.065406996 | Down | glycosyl hydrolase (2391 nt) |
| MGG_08774 | -3.790912355 | -1.064388508 | Down | chitin deacetylase (1107 nt) |
| MGG_01208 | -1.560037961 | -1.062644511 | Down | conserved hypothetical protein (639 nt) |
| MGG_01653 | -1.978306305 | -1.056904511 | Down | nuclear transcription factor Y subunit B-3 (615 nt) |
| MGG_13944 | -3.269625202 | -1.054479897 | Down | hypothetical protein (288 nt) |
| MGG_07734 | -2.383553079 | -1.053901938 | Down | M-phase inducer phosphatase (1710 nt) |
| MGG_09949 | -2.024300402 | -1.046379935 | Down | oxysterol-binding protein (1572 nt) |
| MGG_03649 | -1.215690451 | -1.043931126 | Down | protein phosphatase 1 regulatory subunit SDS22 (1146 nt) |
| MGG_09199 | -1.672048117 | -1.042885813 | Down | repressible acid phosphatase (1374 nt) |
| MGG_02423 | -1.010896389 | -1.03922131 | Down | ER lumen protein retaining receptor 2 (645 nt) |
| MGG_03219 | -1.467366269 | -1.038280996 | Down | conidiophore development protein hymA (963 nt) |
| MGG_00203 | -1.832486716 | -1.037786759 | Down | conserved hypothetical protein (1881 nt) |
| MGG_01390 | -1.164289467 | -1.031395196 | Down | MFS hexose transporter (1533 nt) |
| MGG_00593 | -3.36099766 | -1.029202535 | Down | liver carboxylesterase (2088 nt) |
| MGG_07569 | -1.330902164 | -1.022779869 | Down | cellobiose dehydrogenase (1728 nt) |
| MGG_08201 | -4.122816385 | -1.018223292 | Down | DUF636 domain-containing protein (411 nt) |
| MGG_07872 | -1.06287969 | -1.017921908 | Down | conserved hypothetical protein (477 nt) |
| MGG_12671 | -1.353471223 | -1.016753905 | Down | DUF718 domain-containing protein (447 nt) |
| MGG_02982 | -1.055602066 | -1.011866587 | Down | averantin oxidoreductase (1569 nt) |
| MGG_08550 | -1.226015216 | -1.005932956 | Down | lipolytic enzyme (1083 nt) |
| MGG_00631 | -1.991577141 | -1.005719087 | Down | conserved hypothetical protein (1851 nt) |
| MGG_09816 | -2.674503854 | -1.004576686 | Down | conserved hypothetical protein (933 nt) |
新窗口打开
2.5 qRT-PCR验证
为了验证RNA-seq数据的准确性,随机选择了10个DEGs进行qRT-PCR验证(表4)。结果显示,与RNA-Seq数据一致,与在野生菌株Guy11中相比,MGG_01391、MGG_03825、MGG_09000等基因表达水平在Δznf1 和Δpmk1中显著下调,而MGG_08944、MGG_11084在Δznf1 中表达水平显著升高。MGG_02246在Δznf1中下调表达,但在Δpmk1中上调表达26倍(图4)。Table 4
表4
表4用于qRT-PCR验证的基因及其RNA-Seq测序结果
Table 4The RNA-Seq result of genes selected for qRT-PCR analysis
| 基因编号 Gene ID | 描述 Description | Log2 ratio (Δznf1/Guy11) | Log2 ratio (Δpmk1/Guy11) |
|---|---|---|---|
| MGG_10422 | C6转录因子OefC C6 transcription factor OefC | -1.6 | -6.6 |
| MGG_01391 | 贝壳杉烯氧化酶Ent-kaurene oxidase | -3.0 | -5.3 |
| MGG_03825 | Isotrichodermin C-15羟化酶 Isotrichodermin C-15 hydroxylase | -3.3 | -9.8 |
| MGG_09000 | 蛋白激酶结构域 Protein kinase domain-containing protein | -2.2 | -11.7 |
| MGG_09994 | 脂质磷酸磷酸酶1 Lipid phosphate phosphatase 1 | -3.2 | -2.9 |
| MGG_05719 | 30 kD热激蛋白 30 kD heat shock protein | -2.1 | -3.4 |
| MGG_03329 | 小分子热激蛋白 Small heat shock protein | -3.2 | -2.6 |
| MGG_02246 | 保守假定蛋白Conserved hypothetical protein | -1.7 | 3.9 |
| MGG_08944 | 预测蛋白 Predicted protein | 6.1 | 0.10 |
| MGG_11084 | TPR结构域蛋白 TPR domain-containing protein | 5.1 | -0.64 |
新窗口打开
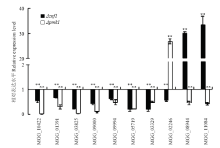 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图410个差异表达基因的qRT-PCR分析纵轴表示与在野生菌株Guy11中相比(定为1),10个DEGs在Δznf1 或Δpmk1中相对表达水平,“**”代表与Guy11相比,差异极显著(P<0.01),误差线表示标准差
-->Fig. 4qRT-PCR analysis on 10 DEGsThe vertical axis represented the relative expression levels of the 10 DEGs in Δznf1 or Δpmk1 when compared with the wild-type strain Guy11 (arbitrarily set to 1). Asterisks indicated significant differences (**, P<0.01). Error bars represented standard deviation
-->
3 讨论
RNA-Seq是用来研究某一生物对象在特定生物过程中基因表达差异的技术[39-40]。该技术结合了转录组建库实验方法与数字基因表达谱的信息分析手段,可广泛应用于生理调控、农业性状、环境改造等领域。该技术主要用来研究基因表达的情况,在基因表达量分析、基因表达差异分析以及差异基因模式和功能聚类等方面应用广泛[41]。近年来,基于RNA-Seq的全基因表达谱分析技术已广泛应用于稻瘟病菌,为稻瘟病菌基因功能和结构的研究提供了许多有参考价值的数据。SOANES等[42]通过RNA-Seq和HT-SuperSAGE技术研究了稻瘟病菌在不同营养生长条件以及附着胞形成各阶段中基因的差异表达情况,鉴定到了一组在附着胞形成阶段特异高表达的基因;LI等[43]利用RNA-Seq初步明确了稻瘟病菌产孢相关转录因子Cos1的调控基因;PHAM等[44]将Chip-Seq和RNA-Seq数据结合分析了组蛋白H3K4甲基转移酶MoSET1在营养生长阶段和附着胞发育阶段的调控网络。稻瘟病菌Znf1是与附着胞形成、侵染、致病过程相关的转录因子[18],利用RNA-Seq技术,可为进一步鉴定Znf1下游调控基因,明确Znf1的调控网络提供参考数据。因此,本研究中,笔者对稻瘟病菌野生菌株Guy11和突变体Δznf1 进行RNA-Seq分析,获得了Δznf1在营养生长阶段的差异表达基因。同时,对稻瘟病菌附着胞形成调控因子PMK1的缺失突变体也进行了RNA-Seq测序,并筛选出了Δznf1和Δpmk1中共同的差异表达基因。测序质量评估分析显示测序质量较高,qRT-PCR验证表明测序结果可靠,测序所得数据可为后续研究提供依据。Δznf1表达谱分析共筛选到709个差异表达基因,其中上调表达基因299个,下调表达基因410个(附表1)。本试验差异表达基因的筛选标准为∣log2 ratio (Δznf1/Guy11)∣≥1,且FDR≤0.001,因此可能还有部分受Znf1调控的基因,但不符合标准而未被筛选到。一些与稻瘟病菌致病相关的基因表达与Znf1有关。例如,HOX7表达水平在Δznf1 中有一定程度的下调(表3),且Δhox7与Δznf1 的表型类似,均不能形成成熟的附着胞,仅在芽管或菌丝尖端形成无黑色素化的膨大结构[16],因此,HOX7可能直接或间接受Znf1调控。之前的研究发现,与野生菌株Guy11相比,Δznf1对胞壁降解酶Glucanex敏感性升高,同等条件下Δznf1原生质体释放量增加[18]。用等量的Glucanex对Guy11和Δznf1菌丝处理2.5 h,计数显示,Guy11原生质体释放量为(15.2±1.17)×107个,而Δznf1原生质体释放量为(27.2±0.71)×107个。本试验RNA-Seq数据显示几丁质合成相关基因CHS3以及α-1,3-葡聚糖编码基因AGS1在Δznf1中有一定程度的下调表达(表3)。因此,笔者推测Znf1可能通过调控CHS3和AGS1的表达水平从而影响稻瘟病菌细胞壁结构的完整性。在Δznf1差异表达基因中,有近60%的基因编码预测蛋白(hypothetical protein)或保守的假定蛋白(conserved hypothetical protein)。MGG_08944编码一个假定蛋白,在其他丝状真菌中不能找到其同源蛋白。测序数据显示log2 ratio(Δznf1/Guy11)为6.1,qRT-PCR进一步证明其在Δznf1中上调表达30倍。MGG_11005、MGG_14426等在Δznf1中显著下调(附表1)。在今后的研究中,可以通过基因敲除技术进一步明确这些受Znf1调控的假定蛋白基因的功能。最近,有研究以稻瘟病菌70-15菌株为背景,对47个C2H2转录因子基因进行了敲除,其中MGG_14931(本文的ZNF1)命名为VRF1,并对Δvrf1在饥饿诱导下的菌丝进行了表达谱分析[25]。本研究以Δznf1营养菌丝为材料进行表达谱测序,结果与Δvrf1数据有一定的差异,可能是由于菌株的不同以及对菌丝样品的处理不同。
Pmk1是稻瘟病菌附着胞形成相关基因的重要调控因子[42]。Δznf1和Δpmk1表型类似,都不能形成附着胞,对寄主致病性完全丧失,都能响应外源cAMP的诱导[13,18]。在附着胞发育阶段,ZNF1表达水平在Δpmk1中显著下调[18]。将Δznf1和Δpmk1中差异表达基因比较发现,Δznf1里709个差异表达基因中,有397个(约56%)基因同时也受Pmk1调控,其中,317个基因在Δznf1和Δpmk1 中上调/下调趋势相同(图3,附表3)。该结果可以进一步证明Znf1可能是位于Pmk1下游,参与调控附着胞的形成[18]。3个在Δznf1和Δpmk1中均显著下调的基因,MGG_03825、MGG_02329和MGG_08498,都编码isotrichodermin C-15羟化酶,它们都含有细胞色素P450超家族的保守结构域,其中MGG_03825的表达经qRT-PCR验证(图4,附表3)。拟分枝镰孢(Fusarium sporotrichioides)和小麦赤霉病菌(Fusarium graminearum)中TRI11与这3个基因同源,其编码一个细胞色素P450单加氧酶。研究表明TRI11与单端孢霉烯毒素合成相关,主要负责C-15的羟基化反应[45-46]。这3个基因是否与稻瘟病菌生长、产孢、附着胞形成、致病等过程相关尚不明确,有待进一步研究。此外,Δznf1和Δpmk1中的共同差异基因数仅占Δpmk1中差异基因数目(3 169个)的12.6%,表明Znf1和Pmk1在调控机制上存在很大差异,Pmk1下游还存在其他转录因子负责这些差异表达基因的转录。而且,有一些基因在Δznf1和Δpmk1中上调/下调趋势虽然相同,但上调/下调倍数有很大差异。疏水蛋白基因MPG1在Δpmk1中下调倍数达130倍,但在Δznf1中仅有轻微的下调表达。菌落疏水性测定表明Δpmk1 菌落疏水性丧失,但Δznf1菌落疏水性没有明显的改变(数据未显示)。可见,在这些基因的调控上,Znf1与Pmk1有很大的不同。
SOANES等[42]通过RNA-Seq和HT-SuperSAGE技术对稻瘟病菌菌株Guy11在营养生长(CM培养基)阶段和附着胞形成阶段(分生孢子分别诱导4、6、8、14和 16 h)分别进行了表达谱分析,发现与在营养生长阶段相比,共有1 838个基因的表达水平在附着胞形成阶段显著上调。结合本试验数据分析发现,这些在附着胞发育阶段上调表达的基因中有48个基因在Δznf1 和Δpmk1中均显著下调表达(附表4)。比如,MGG_09000编码CMGC/CDK蛋白激酶,其log2 ratio(Δznf1/Guy11)及log2 ratio(Δpmk1/Guy11)分别为-2.23和-11.72,而在CM及附着胞形成阶段(T4、T6、T8、T14、T16)表达量分别为1.96、40.35、25.67、32.17、33.44、49.80[42](附表)。HOX7(MGG_12865)编码一个含Homeobox结构域的转录因子,参与稻瘟病菌附着胞形成、侵染和致病过程[16],其在稻瘟病菌附着胞形成阶段,特别是在分生孢子诱导4 h后表达水平显著升高(附表)。然而,在突变体Δznf1和Δpmk1中,HOX7表达水平降低。这些与附着胞形成相关的基因同时受Znf1和Pmk1调控,表明与Pmk1类似,Znf1也参与这些附着胞相关基因的调控。
Table S4
附表4
附表4在附着胞形成阶段上调表达但在Δznf1和Δpmk1中下调表达的基因[
Table S4Genes up-regulated during appressorium formation, but down-regulated in Δznf1 and Δpmkl
| 基因编号 Gene ID | Log2 ratio (Δpmk1/Guy11) | Log2 ratio (Δznf1/Guy11) | 描述 Description |
|---|---|---|---|
| MGG_02942 | -1.468094002 | -4.161723188 | conserved hypothetical protein (1749 nt) |
| MGG_05250 | -1.389007222 | -3.245205238 | conserved hypothetical protein (2520 nt) |
| MGG_02290 | -3.545532356 | -3.141142101 | conserved hypothetical protein (858 nt) |
| MGG_01391 | -5.276496666 | -3.070166018 | ent-kaurene oxidase (1569 nt) |
| MGG_10322 | -1.926158373 | -2.87570589 | cortical patch protein (765 nt) |
| MGG_12865 | -1.990940661 | -2.412767326 | conserved hypothetical protein (1344 nt) |
| MGG_01368 | -2.284535475 | -2.345597001 | conserved hypothetical protein (714 nt) |
| MGG_10429 | -1.969063833 | -2.277318632 | conserved hypothetical protein (963 nt) |
| MGG_07088 | -1.983069604 | -2.257691984 | hypothetical protein (315 nt) |
| MGG_09000 | -11.72280753 | -2.230954435 | protein kinase domain-containing protein (2160 nt) |
| MGG_01542 | -1.740912779 | -2.072756342 | endo-1,4-beta-xylanase (996 nt) |
| MGG_15428 | -1.279201609 | -2.012748493 | para-nitrobenzyl esterase (1674 nt) |
| MGG_03764 | -1.207232614 | -2 | salicylate hydroxylase (1395 nt) |
| MGG_09001 | -5.224001674 | -1.915662644 | hypothetical protein (474 nt) |
| MGG_00993 | -3.513254318 | -1.89630787 | vacuolar iron transporter Ccc1 (1014 nt) |
| MGG_09828 | -1.149050919 | -1.763810581 | vacuolar conductance protein (1959 nt) |
| MGG_05358 | -1.222148207 | -1.714428705 | LYR family protein (267 nt) |
| MGG_09323 | -2.623386485 | -1.703160095 | phenylacetone monooxygenase (1944 nt) |
| MGG_02929 | -1.30712216 | -1.675273272 | phosphotransferase enzyme family protein (789 nt) |
| MGG_10359 | -2.127489735 | -1.673771768 | conserved hypothetical protein (1248 nt) |
| MGG_10422 | -6.573787425 | -1.646850887 | C6 transcription factor OefC (2112 nt) |
| MGG_01798 | -1.143330494 | -1.587742596 | conserved hypothetical protein (906 nt) |
| MGG_15228 | -2.91083778 | -1.538419915 | predicted protein (354 nt) |
| MGG_05897 | -1.372405891 | -1.535135392 | predicted protein (462 nt) |
| MGG_15174 | -1.676243008 | -1.476305437 | predicted protein (342 nt) |
| MGG_04594 | -1.06210985 | -1.422470257 | conserved hypothetical protein (1410 nt) |
| MGG_06260 | -3.62058641 | -1.418952549 | conserved hypothetical protein (2727 nt) |
| MGG_07211 | -1.990331352 | -1.418645279 | conserved hypothetical protein (1482 nt) |
| MGG_05871 | -2.888578717 | -1.397592365 | integral membrane protein (1896 nt) |
| MGG_05051 | -1.568568062 | -1.385121921 | predicted protein (864 nt) |
| MGG_05114 | -1.957295549 | -1.380370367 | hypothetical protein (1662 nt) |
| MGG_00899 | -1.038913905 | -1.372926068 | tubulin-specific chaperone C (1056 nt) |
| MGG_04835 | -1.101212251 | -1.367605669 | conserved hypothetical protein (1008 nt) |
| MGG_03392 | -3.572023445 | -1.357010554 | cytochrome P450 11B1, mitochondrial precursor (1527 nt) |
| MGG_01612 | -2.97012947 | -1.320419788 | hypothetical protein (1818 nt) |
| MGG_07455 | -1.525919085 | -1.252739522 | sorbitol utilization protein SOU2 (879 nt) |
| MGG_00819 | -1.540981307 | -1.252476214 | niemann-Pick C1 protein precursor (3588 nt) |
| MGG_10059 | -2.754979047 | -1.236333192 | Gelsolin repeat-containing protein (1281 nt) |
| MGG_10058 | -1.574549094 | -1.195351422 | predicted protein (813 nt) |
| MGG_05976 | -1.212222528 | -1.190497206 | transesterase (1218 nt) |
| MGG_10525 | -1.593134174 | -1.170024378 | conserved hypothetical protein (2520 nt) |
| MGG_09371 | -1.111678983 | -1.145365428 | phosphatidate cytidylyltransferase (1143 nt) |
| MGG_05521 | -1.41214922 | -1.142717043 | conserved hypothetical protein (1707 nt) |
| MGG_08983 | -2.207727034 | -1.121015401 | catechol 1,2-dioxygenase 1 (960 nt) |
| MGG_01653 | -1.978306305 | -1.056904511 | nuclear transcription factor Y subunit B-3 (615 nt) |
| MGG_09199 | -1.672048117 | -1.042885813 | repressible acid phosphatase (1374 nt) |
| MGG_01390 | -1.164289467 | -1.031395196 | MFS hexose transporter (1533 nt) |
| MGG_12671 | -1.353471223 | -1.016753905 | DUF718 domain-containing protein (447 nt) |
新窗口打开
本研究获得的Δznf1在菌丝生长阶段的差异表达基因数目相对较少,而且表达差异倍数在10倍以上的基因不多,可能是由于Δznf1营养生长与野生型Guy11相比没有明显的差异。与野生型菌株相比,Δznf1产孢量略微增多,不能形成附着胞,不能穿透寄主表皮和进行侵染生长,说明病菌的这些发育阶段受Znf1调控,但调控的基因可能未被筛选到。此外,有小部分基因无论是在Guy11还是Δznf1或Δpmk1中表达丰度都较低,这可能与这些基因呈阶段特异性表达有关。因此,在后续的研究中可对Δznf1在分生孢子、附着胞形成的不同阶段、侵染阶段的样品进行RNA-Seq测序,获得差异表达基因,并与本试验的数据比较分析,获得阶段性受Znf1调控的基因,为更系统深入的研究稻瘟病菌致病分子机理提供参考数据。
4 结论
利用RNA-Seq技术获得了Δznf1在营养生长阶段709个差异表达基因的信息,其中一些稻瘟病菌致病相关基因以及附着胞阶段表达水平升高的基因受Znf1调控;397个基因受Znf1和Pmk1共同调控。研究结果为进一步明确稻瘟病菌Znf1转录调控机制以及挖掘新的稻瘟病菌致病相关基因提供了数据资源。The authors have declared that no competing interests exist.
