 ,1, 洪艳云1, 刘登望2, 张博文2, 易图永
,1, 洪艳云1, 刘登望2, 张博文2, 易图永 ,1,*, 李林
,1,*, 李林 ,2,*
,2,*Effects of calcium application on the structural diversity of endophytic bacterial community in peanut roots under acidic red soil cultivation
ZHANG Wei ,1, HONG Yan-Yun1, LIU Deng-Wang2, ZHANG Bo-Wen2, YI Tu-Yong
,1, HONG Yan-Yun1, LIU Deng-Wang2, ZHANG Bo-Wen2, YI Tu-Yong ,1,*, LI Lin
,1,*, LI Lin ,2,*
,2,*通讯作者:
收稿日期:2020-04-2接受日期:2020-09-13网络出版日期:2021-01-12
| 基金资助: |
Received:2020-04-2Accepted:2020-09-13Online:2021-01-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (4084KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张玮, 洪艳云, 刘登望, 张博文, 易图永, 李林. 施钙对酸性红壤花生根系内生细菌群落结构的影响[J]. 作物学报, 2021, 47(1): 116-124. doi:10.3724/SP.J.1006.2021.04087
ZHANG Wei, HONG Yan-Yun, LIU Deng-Wang, ZHANG Bo-Wen, YI Tu-Yong, LI Lin.
花生(Arachis hypogaea L.)是一种重要的油料作物, 在我国境内广泛种植[1]。然而我国花生主产区受海洋大气环流影响, 降雨量高, 钙素极易流失, 从而导致土壤酸化缺钙[2]。花生是嗜钙作物, 缺钙会影响果实发育, 形成空壳, 造成减产甚至绝收[3], 严重影响经济效益[4,5,6]。同时缺钙容易诱发籽仁生理病变使之更易感染真菌病害。在植物与病原菌互作过程中, 钙参与植物的防御反应和组织结构的形成, 增强植物细胞稳定性和抗病能力[7]。钙能直接抑制病原菌菌丝生长、孢子萌发、附着胞形成等, 甚至能参与病原真菌的致病性调控[8,9,10,11]。
近期研究表明, 微生物菌群可以影响植物和病原物之间的相互作用, 有益微生物能够促进植物的生长, 同时增加植物对非生物胁迫的耐受性[12]。许多根际促生菌能够增强宿主抵抗病原物的能力, 通过诱导系统抗性(induced systemic resistance, ISR)使地上部分能抵抗病原微生物的侵袭和害虫的取食[13,14]。同时植物根系分泌物也能通过改变土壤理化性质吸引附近土壤环境微生物向根部聚集, 进而影响根际微生物的群落结构[15]。Yuan等[16]证实, 拟南芥在受到叶部病原菌丁香假单胞菌番茄致病变种(Pseudomonas syringae pv. tomato, Pst)侵染后, 可通过增加氨基酸和长链有机酸分泌量等途径改变根系分泌物成分, 从而调控土壤微生物群落来加强自身抵御病害的能力。Molina等[17]通过将恶臭假单胞菌(Pseudomonas putida)、鞘氨醇单胞菌(Sphingomonas sp.)、巴西固氮螺菌(Azospirillum brasilense)以及不动杆菌(Acinetobacter sp.)的混合菌群接种至玉米根部, 从而提高了玉米的抗旱性。Berendsen等[14]用活体营养型卵菌拟南芥无色霜霉菌(Hyaloperonospora arabidopsidis)侵染拟南芥叶片后, 通过促进根际3种特异诱导ISR的细菌增殖, 起到抗霜霉病的作用。这些研究进一步证实了接种微生物混合种比单一菌株在抵御病害与应对胁迫上具有更好的效果。
目前, 植物与微生物互作的研究大多数基于模式生物拟南芥或者水稻、玉米等主要粮食作物, 而对花生根系内生菌群落结构鲜有报道。本研究通过对不同钙处理下花生根系内生菌的群落基因组进行16SrRNA基因V3-V4区测序, 试图探析对钙敏感花生植株在酸性低钙条件下, 施钙处理对其根系微生物群落结构的影响, 为合理施用矿质营养元素、改良酸性土壤品质、降低花生病害发生提供理论依据。
1 材料与方法
1.1 供试材料
试验地点为湖南农业大学耘园科研基地。花生采用国家主推的大籽品种湘花2008, 由湖南农业大学旱地作物研究所选育, 对钙敏感; 酸性低钙土壤取自湖南省浏阳市普迹镇书院村月光坪的第四纪红壤表层土, pH 4.5, 含有机质27.3 g kg-1、全氮(N) 0.82 g kg-1、全磷(P) 0.33 g kg-1、全钾(K) 17 g kg-1、碱解氮(N) 64 mg kg-1、速效钾(K) 82 mg kg-1、阳离子交换量13 cmol (+) kg-1、交换性钙(Ca) 0.74 cmol (1/2 Ca2+) kg-1、交换性镁(Mg) 0.13 cmol (1/2 Mg2+) kg-1、有效锌(Zn) 0.63 mg kg-1、有效硫(S) 143.7 mg kg-1、有效硼(B) 0.12 mg kg-1、有效钼(Mo) 0.08 mg kg-1, 有效磷(P)未检出, 为典型酸性缺钙红壤(依发明专利“花生种质钙敏感性的土壤鉴定方法”进行土壤鉴定, 具体鉴定方法详见专利附录)。选取内径36 cm、高75 cm、壁厚度0.5 cm的PVC管组装成塑料大桶, 一端置于地膜之上以防止供试土壤与试验地原有土壤接触。每桶内装供试土壤90 kg。施钙组每桶施加生石灰(CaO) 12.5 g (125 kg hm-2), 与桶中表层供试土壤充分混匀, 对照组不施加生石灰(CaO)。为保证除钙以外的其他基本养分平衡一致的供给, 本试验除进行施钙及对照处理外, 各样品组其他养分均依临界值进行养分平衡: 于土柱表层0~20 cm处, 参照大田标准(45%氮磷钾等比例复合肥750 kg hm-2)每桶施尿素4.02 g、磷酸二氢钾8.23 g及六水氯化镁8.50 g, 并混匀。施钙组与对照组各相同处理10桶作为重复。依十字将大桶划分为4个区域, 每桶定苗4株(作为不同生育期采集检测样本), 始花期每桶喷施等量硼肥。花生生长发育阶段进行正常的水分和病虫草害的管理。
1.2 试验方法
1.2.1 样品采集及处理 分别于苗期(2019-06- 04)、花针期(2019-06-24)与饱果期(2019-08-14)采集施钙组与对照组植株根系样品。每组设置3个重复。将各组样品植株全株拔出, 抖动根系以去除疏松的大块土壤。用2把无菌剪刀分别在施钙组与对照组样品根茎交界处正下方切下约5 cm的根。抖落大块土壤, 并用灭菌毛刷清除表面小颗粒状土壤, 再用无菌水多次反复冲洗样本表面, 并摘去小侧根。将处理后的根放入5%次氯酸钠溶液浸泡5 min, 再转移至装有适量无菌玻璃珠及无菌水的50 mL灭菌离心管中, 剧烈摇晃以便物理去除根表土壤微生物, 用无菌水重复振荡冲洗3次。称取2 g根系组织, 用灭菌剪刀尽可能剪碎, 放入灭菌三角瓶, 加入20 mL无菌的1×PBS缓冲液, 摇床振荡4 h, 40 KHz超声波超声振荡15 min, 5 μm滤膜真空抽滤。将滤液15,000×g离心15 min, 弃上清收集菌体沉淀。使用EasyPure Genomic DNA Kit试剂盒提取菌体沉淀中的根系内生细菌基因组DNA, 使用NanoDrop2000检测所提样品的核酸质量, 合格后送天津诺禾致源生物信息科技有限公司进行16S扩增子测序, 样品编号如表1所示。Table 1
表1
表116S扩增子测序样品分组编号
Table 1
| 生育期 Stage | 不施钙组送样编号 Sample groups without calcium application | 施钙组送样编号 Sample groups with calcium application |
|---|---|---|
| 苗期 Seeding stage | B.R.1, B.R.2, B.R.3 | BC.R.1, BC.R.2, BC.R.3 |
| 花针期 Acicula forming stage | F.R.1, F.R.2, F.R.3 | FC.R.1, FC.R.2, FC.R.3 |
| 饱果期 Pod maturing stage | P.R.1, P.R.2, P.R.3 | PC.R.1, PC.R.2, PC.R.3 |
新窗口打开|下载CSV
1.2.2 16S扩增子测序 取适量样本DNA于离心管中, 使用无菌水稀释样本至1 ng μL-1, 以稀释后的基因组DNA为模板, 根据测序区域16S V3-V4选择特异引物341F (Forward primer 5°-CCTAYG GGRBGCASCAG-3°)与806R (Reverse primer 5°-GG ACTACNNGGGTATCTAAT-3°)进行PCR。PCR产物使用2%浓度的琼脂糖凝胶进行电泳检测后使用GeneJET试剂盒纯化回收目标条带。使用Ion Plus Fragment Library Kit试剂盒进行文库构建。构建好的文库经质检合格后使用Thermofisher的Ion S5 XL进行上机测序, 对测序后下机的原始数据进行拼接和过滤, 得到Clean Reads。
1.2.3 OTUs聚类分析 默认以97%的一致性(Identity)对Clean Reads进行OTUs (operational Taxonomic units)聚类, 然后通过与数据库Silva132的比对进行物种注释。分析微生物群落结构差异并绘制差异OTUs韦恩图、UPGMA聚类树及属水平下物种相对丰度柱形图。选取属分类水平下Top40优势物种, 使用SPSS软件对施钙处理与对照下不同生育期各物种相对丰度差异进行独立样品T检验统计分析, 显著差异的分析结果使用OriginPro 2019软件绘制柱状图。
1.2.4 LEfSe分析 LEfSe (LDA effect size)是一种用于发现和解释高维度生物标识(基因、通路和分类单元)的分析工具, 它强调统计意义和生物相关性, 能够在组与组之间寻找具有统计学差异的Biomarker, 让研究人员能够识别不同丰度的特征以及相关联的类别, 以便找出哪些核心菌群引起组间群落的显著差异。本研究以3作为LDA Score的筛选值对各样品组测序数据进行LEfSe分析并绘制进化分支图。在进化分支图中, 由内至外辐射的圆圈代表了由门至属(或种)的分类级别。在不同分类级别上的每一个小圆圈代表该水平下的一个分类, 小圆圈直径大小与相对丰度大小呈正比。着色原则为无显著差异的物种统一着色为黄色, 差异物种Biomarker跟随组进行着色。
1.2.5 共发生网络构建 分别对施钙组与对照组样品中相对丰度排名Top100的微生物属进行Pearson相关系数计算, 以优势微生物属作为核心节点, 选取Pearson相关系数大于0.6或小于-0.6且显著差异(P<0.05)的连接作为有效连接, 使用Cytoscape软件进行共发生网络图绘制, 以便更加清晰的看出不同样品组中某个微生物属和其他微生物的关联状况。
2 结果与分析
2.1 根系微生物基因组16S V3-V4的PCR扩增结果
以稀释后基因组DNA为模板, 选择16S V3-V4的特异引物341F (5°-CCTAYGGGRBGCASCAG-3°)与806R (5°-GGACTACNNGGGTATCTAAT-3°)进行PCR扩增。部分样品基因组特异引物扩增后电泳检测结果如图1所示。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1根系内生菌基因组16S V3-V4的PCR扩增电泳图
样品编号同
Fig. 1PCR electrophoresis detection of 16S V3-V4 from endophytic bacteria genome
Sample groups are the same as those given in
2.2 16S扩增子测序数据分析
2.2.1 OTU聚类及物种注释概况 经过物种注释及对不同分类层级统计分析发现, 共有2378个OTUs, 其中能够注释到数据库的OTUs数目为2372 (99.75%), 在门水平上, 发现占据主导地位的物种包括厚壁菌门(Firmicutes)、变形菌门(Proteobacteria)、蓝藻门(Cyanobacteria)、拟杆菌门(Bacteroidetes)及放线菌门(Actinobacteria); 在纲水平上主要有芽孢杆菌纲(Bacilli)、γ-变形菌纲(Gammaproteobacteria)、梭菌纲(Clostridia)、α-变形菌纲(Alphaproteobacteria)及拟杆菌纲(Bacteroidia); 在目水平上的优势物种为芽孢杆菌目(Bacillales)、肠杆菌目(Enterobacteriales)、根瘤菌目(Rhizobiales)、梭菌目(Clostridiales)以及黄单胞菌目(Xanthomonadales); 在科水平上的主要物种为芽孢杆菌科(Bacillaceae)、肠杆菌科(Enterobacteriaceae)、伯克氏菌科(Burkholderiaceae)、根瘤菌科(Rhizobiaceae)及黄单胞菌科(Xanthomonadaceae)。2.2.2 花生根系内生菌群落结构 为分析不同生育期花生根系内生细菌的群落结构, 研究不同样本组之间的相似性, 本研究通过对样本以Binary Jaccard距离矩阵进行UPGMA等级聚类, 构建样本的聚类树。同时根据聚类得到的OTUs分析不同样本组之间共有及特有的OTUs并绘制成韦恩图, 如图2所示。根据差异OTUs韦恩图分析可以发现, 除43个共有的核心OTUs以外, 随着花生植株的生长发育, 各生育期特有的OTUs逐渐减少: 苗期施钙与不施钙样品组特有的OTUs数目为177、117; 花针期施钙组特有24个差异OTUs, 而不施钙组特有47个差异OTUs; 饱果期施钙组与对照组的差异OTUs数分别为5、8。
图2
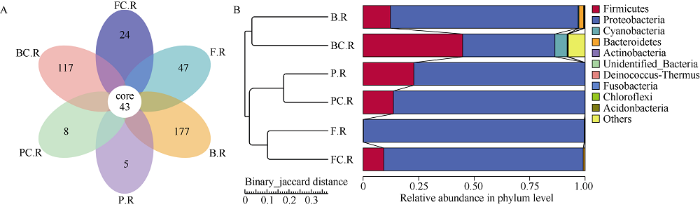 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2Venn分析与UPGMA聚类
A: 不同样品组根系内生菌差异OTUs韦恩图; B: 不同样品组根系内生菌UPGMA聚类树。样品编号同
Fig. 2Venn analysis and UPGMA clustering
A: Venn diagram of OTUs from the root endophytes in different sample groups; B: UPGMA clustering tree of the root endophytes in different sample groups. Sample groups are the same as those given in
选取不同生育期不同处理下各样品组在属(Genus)分类水平上相对丰度排名前20的物种绘制物种相对丰度柱形图, 以便直观查看各样品组中相对丰度较高的优势物种及其所占比例(图3)。根据测序结果, 统计了各生育期不同处理样品组的优势物种(相对丰度排名前5的优势属), 如表2所示。苗期不施钙样品组优势菌属有不动杆菌属(Acinetobacter: 16.9%)、克雷伯氏菌属(Klebsiella: 16.8%)、肠杆菌属(Enterobacter: 7.5%)、厌氧芽孢杆菌属(Anoxybacillus: 5.0%)、草螺菌属(Herbaspirillum: 4.9%)等; 苗期施钙样品组优势菌属包括克雷伯氏菌属(Klebsiella: 30.1%)、芽孢杆菌属(Bacillus: 26.3%)、肠杆菌属(Enterobacter: 6.1%)、水栖菌属(Enhydrobacter: 5.1%)、慢生根瘤菌属(Bradyrhizobium: 1.8%)等; 花针期不施钙样品组优势菌属有克雷伯氏菌属(Klebsiella: 16.9%)、肠杆菌属(Enterobacter: 8.9%)、不动杆菌属(Acinetobacter: 6.7%)、泛菌属(Pantoea: 5.0%)、戴氏菌属(Dyella: 3.7%)等; 花针期施钙样品组优势菌属有克雷伯氏菌属(Klebsiella: 37.2%)、寡养单胞菌属(Stenotrophomonas: 13.1%)、假单胞菌属(Pseudomonas: 8.7%)、从毛单胞菌属(Comamonas: 8.3%)、肠杆菌属(Enterobacter: 7.4%)等; 饱果期不施钙样品组优势菌属为克雷伯氏菌属(Klebsiella: 33.4%)、肠杆菌属(Enterobacter: 11.3%)、类芽孢杆菌属(Paenibacillus: 6.3%)、假单胞菌属(Pseudomonas: 4.9%)、芽孢杆菌属(Bacillus: 1.7%)等; 饱果期施钙样品组优势菌属为克雷伯氏菌属(Klebsiella: 30.9%)假单胞菌属(Pseudomonas: 8.9%)、类芽孢杆菌属(Paenibacillus: 8.7%)、赖氨酸芽孢杆菌属(Lysinibacillus: 8.3%)、芽孢杆菌属(Bacillus: 3.2%)等; 部分主要优势物种未鉴定到属分类水平, 如伯克氏菌科(Burkholderiaceae)、根瘤菌科(Rhizobiaceae)、梭菌目(Clostridiales)、蓝藻门(Cyanobacteria)等故而未列入上述不同样品组中优势物种排名。综合各样品组数据发现, 克雷伯氏菌属(Klebsiella)、肠杆菌属(Enterobacter)在所有样品组中均为优势物种且相对丰度较高。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3不同样品组花生根系内生菌Top 20物种相对丰度柱形图
样品编号同
Fig. 3Histogram of the Top 20 genera relative abundance from different sample groups
Sample groups are the same as those given in
Table 2
表2
表2属分类水平下不同生育期不同处理相对丰度百分比Top 10的根系内生菌
Table 2
| 处理 Treatment | 样品编号 Sample group | 优势菌属 Dominant genus | 相对丰度百分比PRA (%) | 处理 Treatment | 样品编号 Sample group | 优势菌属 Dominant genus | 相对丰度百分比PRA(%) |
|---|---|---|---|---|---|---|---|
| 苗期不施钙 Seeding stage without calcium | B.R. | Acinetobacter | 16.9 | 苗期施钙 Seeding stage with calcium | BC.R. | Klebsiella | 30.1 |
| Klebsiella | 16.8 | Bacillus | 26.3 | ||||
| Enterobacter | 7.5 | Enterobacter | 6.1 | ||||
| Anoxybacillus | 5.0 | Enhydrobacter | 5.1 | ||||
| Herbaspirillum | 4.9 | Bradyrhizobium | 1.8 | ||||
| 花针期不施钙 Acicula forming stage without calcium | F.R. | Klebsiella | 16.9 | 花针期施钙 Acicula forming stage with calcium | FC.R. | Klebsiella | 37.2 |
| Enterobacter | 8.9 | Stenotrophomonas | 13.1 | ||||
| Acinetobacter | 6.7 | Pseudomonas | 8.7 | ||||
| Pantoea | 5.0 | Comamonas | 8.3 | ||||
| Dyella | 3.7 | Enterobacter | 7.4 | ||||
| 饱果期不施钙 Pod maturing stage without calcium | P.R. | Klebsiella | 33.4 | 饱果期施钙 Pod maturing stage with calcium | PC.R. | Klebsiella | 30.9 |
| Enterobacter | 11.3 | Pseudomonas | 8.9 | ||||
| Paenibacillus | 6.3 | Paenibacillus | 8.7 | ||||
| Pseudomonas | 4.9 | Lysinibacillus | 8.3 | ||||
| Bacillus | 1.7 | Bacillus | 3.2 |
新窗口打开|下载CSV
2.2.3 优势菌属差异 为了寻找不同处理下不同生育期样品组中具有显著差异的微生物属, 选取属分类水平下Top40优势物种, 对施钙处理与对照下的不同生育期各物种相对丰度差异进行独立样品T检验(T-test)统计分析(图4)发现, 施钙组与对照组样品在苗期与饱果期时, 虽然部分微生物属在相对丰度上存在差异, 但并不显著(P>0.05), 而花针期施钙处理与对照之间, 假单胞菌属(Pseudomonas: P=0.025)、赖氨酸芽孢杆菌属(Lysinibacillus: P=0.037)及劳尔氏菌属(Ralstonia: P=0.038)的相对丰度存在显著差异(P≤0.05)。其中假单胞菌属(Pseudomonas)与赖氨酸芽孢杆菌属(Lysinibacillus)施钙组中的相对丰度较不施钙组明显增加而劳尔氏菌属(Ralstonia)的相对丰度则在施钙组中显著降低。前人研究发现假单胞菌属(Pseudomonas)与赖氨酸芽孢杆菌属(Lysinibacillus)的部分菌株具有抑菌活性, 降解有害物质甚至抵御病害侵袭的作用, 而劳尔氏菌属(Ralstonia)与花生青枯病病原菌在分类水平上为同一属。施钙对这些优势微生物属相对丰度的显著影响极有可能与这些菌属的生物学功能息息相关。施钙可能在一定程度上能抑制某些病原菌生长, 从而有利于花生抵御相关病害。
图4
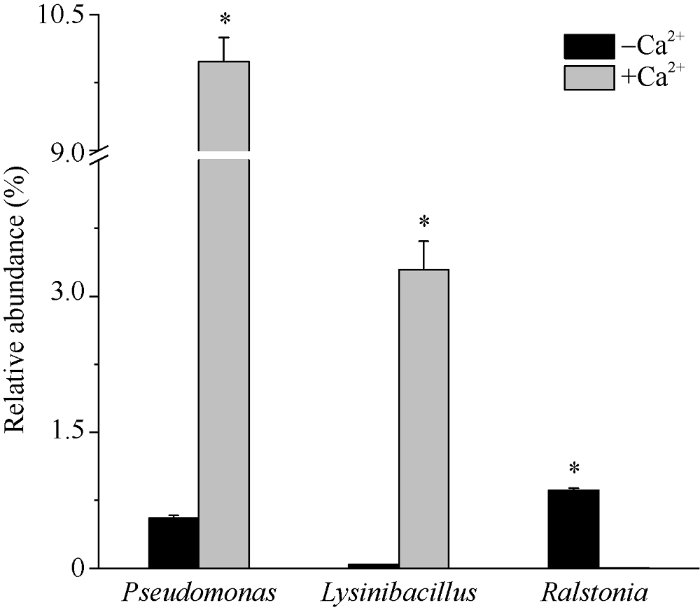 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4花针期相对丰度差异显著微生物属
Fig. 4Genera with significant differences in relative abundance at acicula forming stage
LEfSe统计分析结果如图5 (LEfSe进化分支图)所示, 红色节点表示在苗期不施钙样品组中起到重要作用的微生物类群, 绿色节点表示在苗期施钙样品组中起到重要作用的微生物类群, 蓝色节点表示在花针期不施钙样品组中起到重要作用的微生物类群, 紫色节点表示在花针期施钙样品组中起到重要作用的微生物类群, 浅蓝色节点表示在饱果期不施钙样品组中起到重要作用的微生物类群, 橙色节点表示在饱果期施钙样品组中起到重要作用的微生物类群。进化分支图结果显示, 紫色扇形区域与橙色扇形区域较大且相关节点较多, 表明花针期与饱果期施钙样品组中能起到关键生物学作用的微生物类群较多。属分类水平上: 施钙样品组中苗期的慢生根瘤菌属(Bradyrhizobium), 花针期的希瓦氏菌属(Shewanella)、假单胞菌属(Pseudomonas)及寡养单胞菌属(Stenotrophomonas), 饱果期的类芽孢杆菌属(Paenibacillus)、赖氨酸芽孢杆菌属(Lysinibacillus)与短波单胞菌属(Brevundimonas)等优势物种分别在不同的生育期起关键性生物学作用。
图5
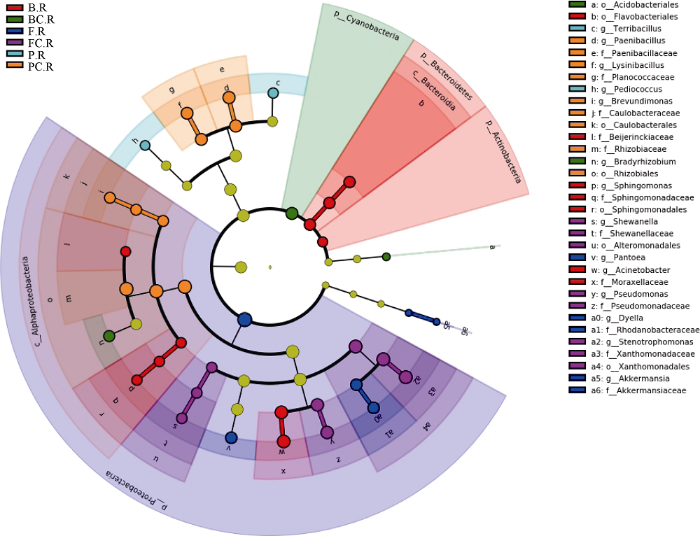 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5不同样品组花生根系内生菌LEfSe分析进化分支图
样品编号同
Fig. 5LEfSe analysis cladogram of endophytic bacteria from different sample groups
Sample groups are the same as those given in
2.2.4 Network共发生网络分析 网络分析被广泛应用于复杂微生物群落的探究中。为进一步探究不同处理下花生根系内生细菌间的相互作用, 本研究计算差异菌属两两之间pearson相关系数并构建了共发生网络(图6)。在施钙组微生物菌属共发生网络中, 核心菌属类诺卡式菌属(Nocardioides)与18个菌属显著正相关, 溶杆菌属(Lysobacter)和中生根瘤菌属(Mesorhizobium)均与17个菌属显著正相关, 细链孢菌属(Catenulispora)与16个菌属显著正相关。而相应的在不施钙处理下样品组根系内生菌关联互作网络中, 发现核心菌属潘多拉菌属(Pandoraea)与其他菌属之间的总关联系数最高, 而戴氏菌属(Dyella)与16个菌属显著正相关。丛毛单胞菌属(Comamonas)、罗河杆菌属(Rhodanobacter)、芽孢杆菌属(Bacillus)、劳尔氏菌属(Ralstonia)等优势菌属也与其他菌属之间存在不同程度的关联互作。推测在施钙处理的花生根系内生细菌群落中, 类诺卡式菌属(Nocardioides)、溶杆菌属(Lysobacter)、中生根瘤菌属(Mesorhizobium)以及细链孢菌属(Catenulispora)作为核心菌属具有重要调控作用, 且对网络内其他微生物种群影响较大。而在不施钙处理的花生根系内生细菌群落中, 潘多拉菌属(Pandoraea)、戴氏菌属(Dyella)、丛毛单胞菌属(Comamonas)、罗河杆菌属(Rhodanobacter)、芽孢杆菌属(Bacillus)、劳尔氏菌属(Ralstonia)等可能为该微生物群落的核心调控菌属, 对其他微生物种群具有较大的影响。同时可以明显看出, 以类诺卡式菌属(Nocardioides)为核心的施钙组菌群之间互作联系更为紧密, 有效关联更多。表明施钙能在一定程度上增强根系内生菌群之间的互作联系, 使相互关联紧密。
图6
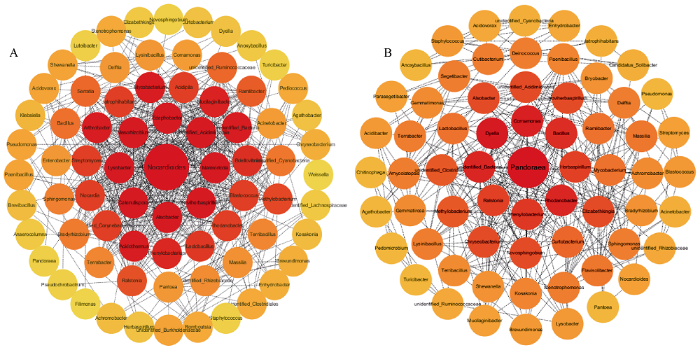 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不同处理下根系内生菌共发生网络图
A: 施钙样品组根系内生菌关联互作网络图; B: 不施钙样品组根系内生菌关联互作网络图。
Fig. 6Interaction network of endophytic bacteria under different treatments
A: interaction network of endophytic bacteria with calcium application; B: interaction network of endophytic bacteria without calcium application.
3 讨论
植物根系是植物-微生物互作的重要生境场所。在此微生态区域, 微生物更易受到植物种类、土壤类型、植物生长阶段等多因素的影响。本研究采用高通量测序技术, 摒弃了繁琐复杂的分离纯化菌株, 以新的视角全面探究, 比较分析了施钙处理与对照之间花生根系内生菌群落结构的显著差异: 差异OTUs分析发现, 苗期与饱果期施钙样品组中所含根系内生细菌差异OTUs数显著高于不施钙样品, 即这2个不同生育期施钙花生样品中根系内生细菌多样性较对照组更为丰富。优势菌属差异分析发现, 假单胞菌属(Pseudomonas)与赖氨酸芽孢杆菌属(Lysinibacillus)的相对丰度在施钙组中显著增加, 而劳尔氏菌属(Ralstonia)相对丰度显著降低。克雷伯氏菌属(Klebsiella)、肠杆菌属(Enterobacter)、类芽孢杆菌属(Paenibacillus)、寡养单胞菌属(Stenotrophomonas)等优势物种作为施钙与对照组间显著差异菌群分别在不同的生育期施钙组样品中起关键性生物学作用。同时, 共发生网络的构建展示出施钙能在一定程度上使根系内生细菌种群之间联系更为紧密, 增强优势菌群之间的关联互作; 同时分析结果还表明, 类诺卡式菌属(Nocardioides)、溶杆菌属(Lysobacter)、细链孢菌属(Catenulispora)等是施钙组根系内生菌群落中的核心菌属。Ibanez等[18]的研究发现, 克雷伯氏菌属(Klebsiella)能在花生根系结瘤固氮和促进植株生长, Sharm等[19]发现, 克雷伯氏菌MBE02 (Klebsiella MBE02)能诱导花生产生ISR同时抵御真菌病害。Gong等[20]的研究论述了阿氏肠杆菌Vt-7 (Enterobacter asburiae Vt-7)对黄曲霉的抑菌活性及对黄曲霉毒素的减毒作用, 与此同时Yang等[21]分离的荧光假单胞菌3JW1 (Pseudomonas fluorescens 3JW1)、Wang等[22]发现的青岛假单胞菌新种(Pseudomonas qingdaonensis sp. nov.)、Wang等[23]首次报道的解木糖赖氨酸芽孢杆菌(Lysinibacillus xylanilyticus)等菌株同样具有抑制黄曲霉菌活性, 降解黄曲霉毒素或者产生黄曲霉毒素抑制剂的作用。Haggag等[24]研究证实, 多粘类芽孢杆菌(Paenibacillus polymyxa)能有效生物防治花生冠腐病。魏兰芳等[25]发现, 抗生素溶杆菌13-1 (Lysobacter antibioticus 13-1)菌株产生的抗菌物质能对水稻白叶枯病具有防治效果。类诺卡式菌属(Nocardioides)大多能降解有毒物质, 对石油污染土壤进行生物修复, 雒晓芳等[26]却分离鉴定并探究了白色类诺卡氏菌(Nocardioides albus)对部分病原菌的抑菌活性。云天艳等[27]还从木薯根际分离到能拮抗尖孢镰刀菌的专化型放线菌MS13, 鉴定后属于细链孢菌属(Catenulispora)。
结合这些以往的研究成果可以合理推测, 克雷伯氏菌属(Klebsiella)作为所有样品组中的优势物种在某种程度上具有结瘤固氮促进植株生长以及提高花生产量的作用。施钙能显著增加假单胞菌属(Pseudomonas)与赖氨酸芽孢杆菌属(Lysinibacillus)在根系内生菌群落中的相对丰度, 联合肠杆菌属(Enterobacter)、类芽孢杆菌属(Paenibacillus)、寡养单胞菌属(Stenotrophomonas)等优势物种能在一定条件下抑制黄曲霉活性同时缓解黄曲霉毒素的毒性甚至降低花生冠腐病的发生概率。与对照组相比, 相对丰度显著降低的劳尔氏菌属(Ralstonia)则间接说明施钙极有可能同时抑制花生青枯病的发生。而类诺卡式菌属(Nocardioides)、溶杆菌属(Lysobacter)、细链孢菌属(Catenulispora)等作为施钙组中的核心菌属能对部分病原菌产生抑菌活性。
4 结论
施钙能丰富根系内生细菌群落的多样性, 使群落间更为紧密的关联互作; 也能使根系微生物组具有更好的促生作用, 使植株能更好地抵御病害侵袭; 施钙能增强特定物种的相对丰度从而促进花生植株的生长发育, 提高产量与品质同时可能抑制常见病害如花生冠腐病、花生青枯病的发生, 抑制黄曲霉菌活性甚至降解黄曲霉毒素; 这些研究结果在一定程度上为南方酸性红壤条件下种植花生提供了施肥管理及病害防控等方面的合理模式, 同时为如何改良酸性土壤品质及提高花生产量与品质奠定了理论基础。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/s41588-019-0402-2URLPMID:31043757 [本文引用: 1]

High oil and protein content make tetraploid peanut a leading oil and food legume. Here we report a high-quality peanut genome sequence, comprising 2.54 Gb with 20 pseudomolecules and 83,709 protein-coding gene models. We characterize gene functional groups implicated in seed size evolution, seed oil content, disease resistance and symbiotic nitrogen fixation. The peanut B subgenome has more genes and general expression dominance, temporally associated with long-terminal-repeat expansion in the A subgenome that also raises questions about the A-genome progenitor. The polyploid genome provided insights into the evolution of Arachis hypogaea and other legume chromosomes. Resequencing of 52 accessions suggests that independent domestications formed peanut ecotypes. Whereas 0.42-0.47 million years ago (Ma) polyploidy constrained genetic variation, the peanut genome sequence aids mapping and candidate-gene discovery for traits such as seed size and color, foliar disease resistance and others, also providing a cornerstone for functional genomics and peanut improvement.
[本文引用: 1]
[本文引用: 1]
URLPMID:28769950 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.plantsci.2018.04.012URLPMID:30709488 [本文引用: 1]

Ca(2+) is a universal second messenger in many signaling pathways in all eukaryotes including plants. Transient changes in [Ca(2+)]cyt are rapidly generated upon a diverse range of stimuli such as drought, heat, wounding, and biotic stresses (infection by pathogenic and symbiotic microorganisms), as well as developmental cues. It has been known for a while that [Ca(2+)]cyt transient signals play crucial roles to activate plant immunity and recently significant progresses have been made in this research field. However the identity and regulation of ion channels that are involved in defense related Ca(2+) signals are still enigmatic. Members of two ligand gated ion channel families, glutamate receptor-like channels (GLRs) and cyclic nucleotide-gated channels (CNGCs) have been implicated in immune responses; nevertheless more precise data to understand their direct involvement in the creation of Ca(2+) signals during immune responses is necessary. Furthermore, the study of other ion channel groups is also required to understand the whole picture of the intra- and inter-cellular Ca(2+) signalling network. In this review we summarize Ca(2+) signals in plant immunity from an ion channel point of view and discuss future challenges in this exciting research field.
DOI:10.1038/nature22009URLPMID:28300100 [本文引用: 1]

Plants encounter a myriad of microorganisms, particularly at the root-soil interface, that can invade with detrimental or beneficial outcomes. Prevalent beneficial associations between plants and microorganisms include those that promote plant growth by facilitating the acquisition of limiting nutrients such as nitrogen and phosphorus. But while promoting such symbiotic relationships, plants must restrict the formation of pathogenic associations. Achieving this balance requires the perception of potential invading microorganisms through the signals that they produce, followed by the activation of either symbiotic responses that promote microbial colonization or immune responses that limit it.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.13343/j.cnki.wsxb.20170073URL [本文引用: 1]

植物病害的微生物防治研究主要集中在植物、病原菌和生防菌三者的互作关系上,相对忽视了植物微生物组/群的作用。越来越多的研究表明,植物内生微生物、根围土壤微生物和叶围微生物均不同程度地参与了植物防病的机制。为了更好地了解相关进展,本文选择部分代表性研究,详述了植物微生物组/群的构成,并结合案例介绍了植物微生物组/群对寄主植物的防/致病作用、对植物病原菌致病性的影响,以及施用生防菌对植物微生物组/群的影响。微生物组学的发展为生防机制领域提出了新的研究思路,有利于发现更加科学的防治手段。
[本文引用: 1]
DOI:10.1038/s41396-018-0093-1URLPMID:29520025 [本文引用: 2]

Disease suppressive soils typically develop after a disease outbreak due to the subsequent assembly of protective microbiota in the rhizosphere. The role of the plant immune system in the assemblage of a protective rhizosphere microbiome is largely unknown. In this study, we demonstrate that Arabidopsis thaliana specifically promotes three bacterial species in the rhizosphere upon foliar defense activation by the downy mildew pathogen Hyaloperonospora arabidopsidis. The promoted bacteria were isolated and found to interact synergistically in biofilm formation in vitro. Although separately these bacteria did not affect the plant significantly, together they induced systemic resistance against downy mildew and promoted growth of the plant. Moreover, we show that the soil-mediated legacy of a primary population of downy mildew infected plants confers enhanced protection against this pathogen in a second population of plants growing in the same soil. Together our results indicate that plants can adjust their root microbiome upon pathogen infection and specifically recruit a group of disease resistance-inducing and growth-promoting beneficial microbes, therewith potentially maximizing the chance of survival of their offspring that will grow in the same soil.
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s40168-018-0537-xURLPMID:30208962 [本文引用: 1]

6) and lower exudation levels for sugars, alcohols, and short-chain organic acids (SCOAs) (C
[本文引用: 1]
DOI:10.1016/j.syapm.2008.10.001URLPMID:19054642 [本文引用: 1]

Several bacterial isolates were recovered from surface-sterilized root nodules of Arachis hypogaea L. (peanut) plants growing in soils from Cordoba, Argentina. The 16S rDNA sequences of seven fast-growing strains were obtained and the phylogenetic analysis showed that these isolates belonged to the Phylum Proteobacteria, Class Gammaproteobacteria, and included Pseudomonas spp., Enterobacter spp., and Klebsiella spp. After storage, these strains became unable to induce nodule formation in Arachis hypogaea L. plants, but they enhanced plant yield. When the isolates were co-inoculated with an infective Bradyrhizobium strain, they were even found colonizing pre-formed nodules. Analysis of symbiotic genes showed that the nifH gene was only detected for the Klebsiella-like isolates and the nodC gene could not be amplified by PCR or be detected by Southern blotting in any of the isolates. The results obtained support the idea that these isolates are opportunistic bacteria able to colonize nodules induced by rhizobia.
DOI:10.1038/s41598-019-40930-xURLPMID:30858512 [本文引用: 1]

A halotolerant rhizobacteria, Klebsiella species (referred to MBE02), was identified that had a growth stimulation effect on peanut. To gain mechanistic insights into how molecular components were reprogrammed during the interaction of MBE02 and peanut roots, we performed deep RNA-sequencing. In total, 1260 genes were differentially expressed: 979 genes were up-regulated, whereas 281 were down-regulated by MBE02 treatment as compared to uninoculated controls. A large component of the differentially regulated genes were related to phytohormone signalling. This included activation of a significant proportion of genes involved in jasmonic acid, ethylene and pathogen-defense signalling, which indicated a role of MBE02 in modulating plant immunity. In vivo and in vitro pathogenesis assays demonstrated that MBE02 treatment indeed provide fitness benefits to peanut against Aspergillus infection under controlled as well as field environment. Further, MBE02 directly reduced the growth of a wide range of fungal pathogens including Aspergillus. We also identified possible molecular components involved in rhizobacteria-mediated plant protection. Our results show the potential of MBE02 as a biocontrol agent in preventing infection against several fungal phytopathogens.
DOI:10.1016/j.foodcont.2019.106718URL [本文引用: 1]
[本文引用: 1]
URLPMID:30798341 [本文引用: 1]
DOI:10.1007/s11274-012-1135-xURLPMID:23108663 [本文引用: 1]

50 % (p 90 %. These results showed a wide distribution of biological control bacteria against AFs in Chinese peanut main producing areas and the selected RAE bacteria could potentially be utilized for the biocontrol of toxicogenic fungi.]]>
DOI:10.1111/j.1365-2672.2007.03611.xURLPMID:18005030 [本文引用: 1]

AIM: To investigate the role of biofilm-forming Paenibacillus polymyxa strains in controlling crown root rot disease. METHODS AND RESULTS: Two plant growth-promoting P. polymyxa strains were isolated from the peanut rhizosphere, from Aspergillus niger-suppressive soils. The strains were tested, under greenhouse and field conditions for inhibition of the crown root rot pathogen of the peanut, as well as for biofilm formation in the peanut rhizosphere. The strains' colonization and biofilm formation were further studied on roots of the model plant Arabidopsis thaliana and with solid surface assays. Their crown root rot inhibition performance was studied in field and pot experiments. The strains' ability to form biofilms in gnotobiotic and soil systems was studied employing scanning electron microscope. CONCLUSION: Both strains were able to suppress the pathogen but the superior biofilm former offers significantly better protection against crown rot. SIGNIFICANCE AND IMPACT OF THE STUDY: The study highlights the importance of efficient rhizosphere colonization and biofilm formation in biocontrol.
URL [本文引用: 1]

【目的】为了明确对水稻白叶枯病具有优质防效的生防细菌——抗生素溶杆菌13-1菌株抗菌物质及对白叶枯病的防治效果。【方法】研究采用高效液相色谱、质谱、13C核磁共振谱、氢核磁共振谱、电喷雾质谱等分析方法。【结果】13-1产生4种抑菌活性组分:6-甲氧基-10-氧基-1-吩嗪醇、吩嗪、吩嗪-1-羧酸及1-羟基-6-甲氧基吩嗪。4种吩嗪类物质对水稻白叶枯病原细菌均具有抑菌活性。田间小区试验表明,菌株13-1发酵液对水稻白叶枯病的防效在60%以上。【结论】研究明确了菌株13-1产生的抗菌物质为吩嗪类物质,这是国内关于吩嗪类物质控制水稻白叶枯病的首次报道。
[本文引用: 1]
DOI:10.11882/j.issn.0254-5071.2015.10.013URL [本文引用: 1]

该研究对白色类诺卡氏菌(Nocardioides albus)进行分离鉴定,并通过温度、紫外、酸碱处理,以绿脓杆菌(Pseudomonas aeruginosa)、恶臭假单胞菌(Pseudomonas putida)、枯草杆菌(Bacillus subtilis)、暗黑微绿链霉菌(Streptomyces atrovirens)为指示菌,用琼脂打孔法进行抑菌实验;用药敏片法进行药敏实验,初步探讨分离菌株的药物敏感性以及抑菌性。结果表明,紫外作用30 min及温度对其抑菌性影响不大;强酸、强碱环境中白色类诺卡氏菌基本没有抑菌活性。白色类诺卡氏菌对抗生素较为敏感,药敏性较高。
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
