 ,*山西省农业科学院谷子研究所 / 特色杂粮种质资源发掘与育种山西省重点实验室, 山西长治 046011
,*山西省农业科学院谷子研究所 / 特色杂粮种质资源发掘与育种山西省重点实验室, 山西长治 046011Identification of PEPC genes from foxtail millet and its response to abiotic stress
ZHAO Jin-Feng**, DU Yan-Wei**, WANG Gao-Hong, LI Yan-Fang, ZHAO Gen-You, WANG Zhen-Hua, WANG Yu-Wen, YU Ai-Li ,*Millet Research Institute, Shanxi Academy of Agricultural Sciences / Shanxi Key Laboratory of Genetic Resources and Breeding in Minor Crops, Changzhi 046011, Shanxi, China
,*Millet Research Institute, Shanxi Academy of Agricultural Sciences / Shanxi Key Laboratory of Genetic Resources and Breeding in Minor Crops, Changzhi 046011, Shanxi, China通讯作者:
第一联系人:
收稿日期:2019-07-24接受日期:2020-01-15网络出版日期:2020-02-14
| 基金资助: |
Received:2019-07-24Accepted:2020-01-15Online:2020-02-14
| Fund supported: |
作者简介 About authors
赵晋锋,E-mail:zhaojfmail@126.com,Tel:0355-2204195.。

摘要
关键词:
Abstract
Keywords:
PDF (5266KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵晋锋, 杜艳伟, 王高鸿, 李颜方, 赵根有, 王振华, 王玉文, 余爱丽. 谷子PEPC基因的鉴定及其对非生物逆境的响应特性[J]. 作物学报, 2020, 46(5): 700-711. doi:10.3724/SP.J.1006.2020.94107
ZHAO Jin-Feng, DU Yan-Wei, WANG Gao-Hong, LI Yan-Fang, ZHAO Gen-You, WANG Zhen-Hua, WANG Yu-Wen, YU Ai-Li.
光合作用是植物生长发育过程中极其重要的化学反应[1]。依据光合作用对无机碳固定过程不同, 植物被划分为C3光合途径、C4循环途径及景天酸代谢(CAM)途径植物[2]。C3途径是最原始最基本的二氧化碳同化途径, 可以合成庶糖、淀粉、脂肪和蛋白质等稳定光合产物, 而C4和CAM途径不能独立合成相对稳定的光合作用产物, 其主要作用是对二氧化碳进行固定、运输、暂时存储[3]。C4循环途径比C3循环途径光合能力大幅提高, 主要是因为C4植物中有特殊的花环型结构, 可以使CO2在Rubisco羧化酶周围高浓度聚集, 大大提高了对水分、氮的充分利用, 即使在高光强照射、高温、干旱和CO2浓度低等逆境环境下, C4植物仍然具有较高的光合速率[4]。磷酸烯醇式丙酮酸羧化酶(phosphoenolpyruvate carboxylase, PEPC)是C4植物光合作用的重要关键酶之一, 在1953年首次从藜科植物菠菜中分离出来[5]。研究表明PEPC是一种细胞质酶, 主要功能是以Mg2+或Mn2+为辅助因子, 催化磷酸烯醇式丙酮酸(phosphoenolpyruvate, PEP)和${HCO^{-}_{3}}$生成草酰乙酸(oxaloacetate, OAA) 和无机磷酸(inorganic phosphate, Pi)的不可逆反应[6]。PEPC是一种变构调节酶, 主要在真菌、藻类、蓝细菌、光合细菌、部分非光合细菌、动物以及植物中广泛存在[7]。目前PEPC基因相继在拟南芥、大豆、水稻、玉米、甘蔗等植物中得到鉴定[8,9,10,11]。高等植物中的PEPC是四聚体三维结构, 由约970个氨基酸(110 kDa)组成, C末端含有PEPC保守功能域[12]。PEPC在植物多种代谢途径中具有广泛生理功能, 主要包括参与光合作用碳同化, 提高油料作物种子脂肪酸含量, 促进种子蛋白质合成, 调节细胞质pH值、根系离子吸收、气孔开放、氨同化以及参与逆境胁迫应答[13]。随着转基因研究的发展, 近年来PEPC逆境分子生物学研究逐渐成为新的热点。研究表明PEPC在植物应对逆境胁迫过程中起重要作用[14]。丁在松等[15]研究转PEPC基因水稻在干早、高温、强光等逆境胁迫下的光合特征, 发现逆境胁迫下转基因水稻较对照具有明显光合优势。焦德茂等[16]以转玉米PEPC基因水稻为研究材料, 发现在高温、高光强条件下转基因水稻的光饱和点和饱和光合效率均显著高于对照。方立峰等[17]发现转PEPC基因水稻在苗期干旱胁迫下维持了相对较高的光合能力, 具有较强的抗氧化和渗透调节能力。另外Jeanneau等[18]认为在适度干旱条件下PEPC基因过表达能提高玉米水分利用率, 增加干物质积累量。干旱、盐和冷害等非生物逆境也被报道能够强烈诱导PEPC基因的表达, 参与植物对环境的抗逆反应[19,20,21]。上述研究结果表明逆境胁迫能诱导PEPC基因过量表达, 参与植物体对环境胁迫的抗逆生理生化反应。
谷子[Setaria italica (L.) P. Beauv.]是C4禾本科重要粮食作物和饲草作物, 起源于我国, 在全球一些温带、亚热带和热带的干旱和半干旱地区广泛种植, 具有抗旱、耐瘠、适应性广等特点[22,23,24]。谷子在生长过程中会经常遇到干旱、盐渍、低温、高温、洪涝以及病虫害侵染等, 这些逆境严重影响了谷子的生长发育和产量品质[25]。2012年谷子基因组测序已完成并公布, 为开展谷子抗逆分子生物学研究提供了极为便利的条件[26,27]。迄今为止PEPC研究主要聚焦在C4型PEPC基因在提高C3植物光合作用和耐胁迫应答领域, 在C4作物玉米、高粱中研究较多, 但是在C4作物谷子中PEPC基因非生物逆境相关研究鲜有报道。本研究检索了谷子中的PEPC基因家族成员, 并对其氨基酸序列、蛋白特征、功能、亚细胞定位、启动子区域顺式应答元件等参数特征分析和预测, 随后检测了家族基因在幼苗期逆境胁迫下的动态表达模式及在拔节、抽穗、灌浆3个生育时期干旱胁迫和不同光照处理下的表达情况, 旨在为进一步分析PEPC基因在谷子逆境应答信号途径中的功能和机制以及为利用基因工程方法改善作物光合速率和提高产量提供数据支持。
1 材料与方法
1.1 试验材料
选用谷子品种‘豫谷1号’, 种子保存于山西省农业科学院谷子研究所生物技术课题。在组培室中培养谷子幼苗, 温度为(22±2)℃, 湿度为60%, 光照周期为16 h光照/8 h黑暗。1.2 试验设计
待组培室幼苗长至三叶一心期时, 分别对其进行20% PEG-6000模拟干旱、盐(250 mmol L-1 NaCl)、ABA (100 μmol L-1)和低温(4℃)胁迫处理, 苗期逆境表达谱分析取样时间点为处理后0、1、3、6、12和24 h, 整株取样[28]。旱棚种植‘豫谷1号’, 对照处理生育期内正常浇水; 干旱处理只浇3次关键水, 分别为拔节(6月20日)、抽穗(8月5日)和灌浆期(8月30日)浇透水, 其他时间采用自然控水方式控水。光照处理为当植株出苗后用黑色遮阳网分别遮挡1层(中等光照, 拔节期、抽穗期、灌浆期测量光照强度分别为221.8、411.6、164.6 μmol m-2 s-1), 2层(弱光照, 拔节期、抽穗期、灌浆期测量光照强度分别为52.1、95.8、58.8 μmol m-2 s-1)至成熟收获。光照强度用标智光强测定仪(GM1040)于晴天上午9:00—10:00测量10次数值, 取平均值。干旱处理试验与光照处理试验为同一对照。对照、中等光照处理和弱光照处理生育期内正常浇水, 其他农田管理措施相同。分别在拔节、抽穗、灌浆期时取旱棚干旱处理与光照强度处理材料和对照旗叶叶片, 立即在-80℃冰箱中速冻备用。1.3 谷子PEPC 基因的鉴定及基因结构、蛋白序列分析
在Phytozome数据库中以拟南芥和玉米的已知PEPC氨基酸序列为递交序列, 进行BlastP比对, 搜索具有完整阅读框的谷子同源序列, 获得谷子候选PEPC基因。基因参数和蛋白序列来源于Phytozome数据库。利用ExPASy网站在线工具预测蛋白分子量和等电点。利用PLACE在线软件对基因启动子区域顺式元件进行分析, 采用GSDS软件绘制基因结构图, 用在线工具CELLO v.2.5进行亚细胞定位预测, 预测置信度计算参考Yu等[29]描述方法。使用Blast工具在NCBI上查找不同物种氨基酸同源性序列, 用ClustalX1.83软件分析比对基因序列[30], 用Mega6.0软件邻接法构建不同物种N-J系统进化树[31]。1.4 总RNA提取、cDNA合成及引物设计
按照说明书, 使用生工TRIzol试剂盒提取植物总RNA; 使用生工第1链cDNA合成试剂盒合成cDNA。所用引物由Primer Primer 5.0设计, 引物信息见表1。Table 1
表1
表1本试验所用引物
Table 1
| 基因 Gene | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') |
|---|---|---|
| SiPEPC1 | TGCGTGCTGGAATGAGTTACT | CCAATAAGCATACATCCCGTG |
| SiPEPC2 | CAAGAGCCAAACTATTGACCTG | TCATACCAGCACGCATTTCA |
| SiPEPC3 | ATGGAAGGGTGTCCCAAAGTT | CCAAGAAGAGAACTGAATGAGAGG |
| SiPEPC5 | GCTACTTCGACGACACCATC | AGGAGGAGAACTGGATGAGC |
| SiPEPC6 | GAGGTGGAACTGTGGGAAGA | GTAAAACGTTGCAGGGTTCTAA |
| β-Actin | CAGTGGACGCACAACAGGTAT | AGCAAGGTCAAGACGGAGAAT |
新窗口打开|下载CSV
1.5 Real-time PCR分析
样品cDNA均一化后作为实时定量PCR模板, 以谷子β-actin基因(Seita.7G294000)作为内参基因。反应体系为20 μL, 包含10 μL 2×荧光染料混合液、0.4 μL正向引物(10 μmol L-1)、0.4 μL反向引物(10 μmol L-1)、2 μL cDNA模板、7.2 μL无菌水。经预试验优化后PCR程序为95℃ 3 min; 95℃ 7 s, 57℃ 10 s, 72℃ 15 s, 45个循环。试验设计3次重复, 采用相对定量2-ΔΔCt方法计算基因在某种逆境处理下某个时间点相对于对照的转录水平变化[32]。SiPEPC成员在苗期不同逆境、不同生育期干旱和不同光照下Real-time PCR 表达分析结果利用百迈克云平台在线工具聚类热图绘制(Pretty Heatmaps)热图, 参数默认。2 结果与分析
2.1 谷子PEPC基因成员鉴定与参数分析
通过序列比对和分析, 从谷子中筛选出6个SiPEPC候选基因成员(表2)。根据基因定位在染色体1~9号上的先后位置顺序分别命名为SiPEPC1、SiPEPC2、SiPEPC3、SiPEPC4、SiPEPC5、SiPEPC6。6个基因不均分布在1号、2号、4号、5号染色体上, 其中SiPEPC1、SiPEPC2、SiPEPC3分别分布于1号、2号、4号染色体上, 而SiPEPC4、SiPEPC5、SiPEPC6全部位于5号染色体上。SiPEPC6基因有2种转录方式, 其他5个基因只有1种转录方式。功 能域分析发现, 所有SiPEPC基因都含有PEPC基因特征功能域PEPcase, 功能域在基因中的详细位置见表2。候选SiPEPC蛋白参数分析表明, SiPEPC成员之间基因组长度变化明显, 在4676~8603 bp之间, 编码氨基酸数目在964~1032之间。成员间分子量范围在109.82~114.89 kD之间, 蛋白等电点范围在5.70~7.14之间, 不稳定指数范围在44.10~52.09之间, 脂肪系数范围在85.81~93.87之间, 平均疏水指数范围在-0.418~ -0.277之间。GSDS在线软件分析SiPEPC成员基因结构发现, SiPEPC1、SiPEPC2、SiPEPC3、SiPEPC5、SiPEPC6分别含9、9、8、6、9个内含子, 而SiPEPC4含20个内含子(图1)。Table 2
表2
表2预测谷子PEPC基因参数
Table 2
| 基因 Gene | 基因ID Gene ID | 染色体位置 Chr. location | 基因长度 Genome length (bp) | 氨基酸数目 No. of amino acid | 等电点 pI | 分子量 Molecular weight (kD) | 内含子数 Intron number | 不稳定指数 Instability index | 脂肪系数 Aliphatic index | 平均疏水指数 Grand average of hydropathicity | PEPcse保守域位置 PEPcse domain location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SiPEPC1 | Seita.1G020700 | 1784608-1791259 | 6652 | 965 | 5.83 | 109.82 | 9 | 48.48 | 87.36 | -0.392 | 163-965 |
| SiPEPC2 | Seita.2G173700 | 26198697-26204510 | 5814 | 967 | 5.77 | 110.20 | 9 | 44.74 | 92.41 | -0.351 | 164-967 |
| SiPEPC3 | Seita.4G175200 | 28034663-28043265 | 8603 | 964 | 5.95 | 109.98 | 8 | 45.83 | 85.81 | -0.418 | 162-964 |
| SiPEPC4 | Seita.5G074200 | 6390259-6398514 | 8256 | 1032 | 7.14 | 114.89 | 20 | 52.09 | 93.87 | -0.277 | 144-348, 414-1032 |
| SiPEPC5 | Seita.5G147000 | 13061164-13065839 | 4676 | 1015 | 5.89 | 113.53 | 6 | 49.05 | 87.69 | -0.319 | 213-1015 |
| SiPEPC6 | Seita.5G324500 | 37380598-37387338 | 6741 | 969 | 5.70 | 110.44 | 9 | 44.10 | 89.79 | -0.375 | 165-969 |
新窗口打开|下载CSV
图1
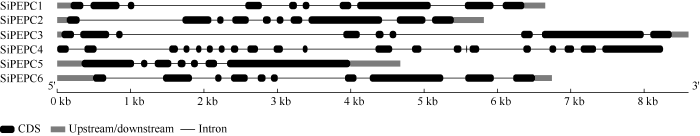 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1SiPEPC成员的基因结构
Fig. 1Gene structure of SiPEPC genes
2.2 SiPEPC蛋白亚细胞定位预测和启动子区域顺式元件分析
采用Psort在线工具对谷子SiPEPC家族成员的亚细胞定位预测表明, SiPEPC成员主要被定位在细胞质、细胞核和线粒体(表3)。利用PLACE在线软件对SiPEPC成员启动子顺式元件分析(表4), SiPEPC成员启动子区域主要包括大量的光应答元件、激素类应答元件(脱落酸、赤霉素、水杨酸、乙烯、茉莉酸甲酯、生长素)、逆境应答元件(逆境防御、低温、干旱、伤害)以及其他生长调控相关顺式元件, 包括缺氧特异性诱导、厌氧诱导必需、分生组织特异性表达、玉米蛋白代谢调控、种子特异性调控、昼夜节律元件等。Table 3
表3
表3SiPEPC蛋白亚细胞定位预测
Table 3
| 基因 Gene | 预测位置 Localization | 可信度 Reliability |
|---|---|---|
| SiPEPC1 | Cytoplasmic; nuclear; mitochondrial | 3.395; 0.685; 0.414 |
| SiPEPC2 | Cytoplasmic; nuclear; mitochondrial | 3.398; 0.831; 0.317 |
| SiPEPC3 | Cytoplasmic; nuclear; mitochondrial | 3.679; 0.620; 0.321 |
| SiPEPC4 | Cytoplasmic; mitochondrial; nuclear | 1.838; 1.290; 1.230 |
| SiPEPC5 | Cytoplasmic; mitochondrial; nuclear | 2.258; 1.092; 0.472 |
| SiPEPC6 | Cytoplasmic; nuclear; mitochondrial | 3.428; 0.758; 0.338 |
新窗口打开|下载CSV
Table 4
表4
表4SiPEPC基因启动子区域顺式元件预测
Table 4
| 顺式元件 cis-element | 典型序列 Typical sequence | 特性 Characteristic | 基因 Gene |
|---|---|---|---|
| Box 4 | ATTAAT | Light responsive element | SiPEPC1, SiPEPC2, SiPEPC3 |
| G-Box | CACGTT | Light responsive element | SiPEPC1, SiPEPC2, SiPEPC3, SiPEPC4, SiPEPC5, SiPEPC6 |
| GATA-motif | AAGGATAAGG | Light responsive element | SiPEPC1, SiPEPC6 |
| AE-box | AGAAACAA | Light responsive element | SiPEPC2, SiPEPC4, SiPEPC6 |
| GT1-motif | GGTTAAT | Light responsive element | SiPEPC2, SiPEPC4, SiPEPC5 |
| ACE | GACACGTATG | Light responsive element | SiPEPC3, SiPEPC5 |
| Sp1 | GGGCGG | Light responsive element | SiPEPC3, SiPEPC4 |
| ABRE | ACGTG | Abscisic acid responsiveness | SiPEPC1, SiPEPC2, SiPEPC3, SiPEPC4, SiPEPC5 |
| P-box | CCTTTTG | Gibberellin-responsive | SiPEPC1, SiPEPC2 |
| TATC-box | TATCCCA | Gibberellin-responsiveness | SiPEPC2, SiPEPC4 |
| GARE-motif | TCTGTTG | Gibberellin-responsive element | SiPEPC6 |
| TCA-element | CCATCTTTTT | Salicylic acid responsiveness | SiPEPC1, SiPEPC2, SiPEPC5, SiPEPC6 |
| ERE | ATTTCATA | Ethylene-responsive element | SiPEPC2, SiPEPC4 |
| CGTCA-motif | CGTCA | MeJA-responsiveness | SiPEPC3, SiPEPC6 |
| TGACG-motif | TGACG | MeJA-responsiveness | SiPEPC3, SiPEPC6 |
| TGA-element | AACGAC | Auxin-responsive element | SiPEPC1, SiPEPC4 |
| TC-rich repeats | GTTTTCTTAC | Defense and stress responsiveness | SiPEPC1, SiPEPC3 |
| LTR | CCGAAA | Low-temperature responsiveness | SiPEPC2, SiPEPC3, SiPEPC4, SiPEPC6 |
| WUN-motif | TTATTACAT | Wound-responsive element | SiPEPC3 |
| MBS | CAACTG | MYB binding site involved in drought-inducibility | SiPEPC5 |
| ARE | AAACCA | Anaerobic induction | SiPEPC1, 3, 4, 6 |
| CCGTCC-box | CCGTCC | Meristem specific activation | SiPEPC1, 2, 4, 6 |
| GC-motif | CCCCCG | Anoxic specific inducibility | SiPEPC1, 2, 6 |
| O2-site | GATGACATGG | Zein metabolism regulation | SiPEPC2, 4, 5 |
| RY-element | CATGCATG | Seed-specific regulation | SiPEPC3 |
| circadian | CAAAGATATC | Circadian control | SiPEPC5 |
| CAT-box | GCCACT | Meristem expression | SiPEPC6 |
新窗口打开|下载CSV
2.3 PEPC序列比对及进化分析
谷子PEPC成员氨基酸序列比对发现成员之间序列非常保守, 相似性较高, 其序列一致性为67.71%。所有成员都含有PEPC蛋白特征功能域PEPcase, 其中SiPEPC1、SiPEPC2、SiPEPC3、SiPEPC5、SiPEPC6序列相似性较高, 整体序列一致性为74.03%, 而SiPEPC4在特征功能域内由于有几个片段的插入和缺失, 因此与其他几个SiPEPC序列一致性较低, 在32.92%~ 34.06%之间。为进一步了解谷子SiPEPC进化关系, 推测其生物功能, 在NCBI数据库中检索了不同物种已知PEPC基因。序列比对发现所有物种PEPC整体一致性非常高, 为72.88%, 说明PEPC基因在进化过程中非常保守。利用 MEGA6.0 软件构建了谷子和不同物种PEPC蛋白成员的Neighbor-Joining进化树(图3)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2PEPC家族蛋白序列的氨基酸序列多重比对
AtPEPC1、AtPEPC2、AtPEPC3: 拟南芥AtPEPC1、AtPEPC2、AtPEPC3; SbPEPC1、SbPEPC2、SbPEPC3: 高粱SbPEPC1、SbPEPC2、SbPEPC3; GmPEPC: 大豆GmPEPC; ZmPEPC1、ZmPEPC2: 玉米ZmPEPC1、ZmPEPC2; StPEPC: 马铃薯StPEPC; AhPEPC: 花生AhPEPC; RcPEPC: 蓖麻RcPEPC; NtPEPC: 烟草 NtPEPC。对应GenBank蛋白序列号分别为Q9MAH0.1、Q5GM68.2、Q84VW9.2、P29195.1、P29194.1、P15804.2、P51061.1、P04711.2、P51059.1、P29196.2、ABY87944.1、ABR29878.1、P27154.1。相同氨基酸残基用黑色表示, 相似氨基酸残基用灰色表示(相似性 ≥ 60%)。
Fig. 2Amino acid sequence alignment of PEPCs
AtPEPC1, AtPEPC2, AtPEPC3: Arabidopsis thaliana AtPEPC1, AtPEPC2, AtPEPC3; SbPEPC1, SbPEPC2, SbPEPC3: Sorghum bicolor SbPEPC1, SbPEPC2, SbPEPC3; GmPEPC: Glycine max GmPEPC; ZmPEPC1, ZmPEPC2: Zea mays ZmPEPC1, 2; StPEPC: Solanum tuberosum StPEPC; AhPEPC: Arachis hypogaea AhPEPC; RcPEPC: Ricinus communis RcPEPC; NtPEPC: Nicotiana tabacum NtPEPC. The corresponding GenBank protein numbers are Q9MAH0.1, Q5GM68.2, Q84VW9.2, P29195.1, P29194.1, P15804.2, P51061.1, P04711.2, P51059.1, P29196.2, ABY87944.1, ABR29878.1, and P27154.1, respectively. The amino acids with an entire homology are shown by a black background, and those shared non-identical conserved identity by a graybackground (≥ 60% similarity).
图3
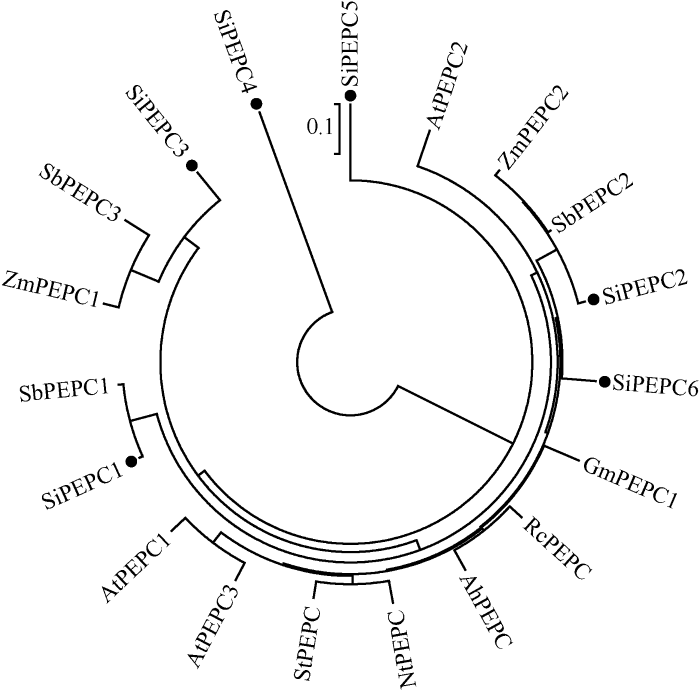 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3不同物种PEPC 蛋白的进化关系
Fig. 3Phylogenetic relationships of PEPC proteins from different species
从图3可以看出, 谷子的SiPEPC基因与高粱、玉米PEPC基因聚合在一起, 揭示谷子与高粱、玉米亲缘关系较近(同属禾本科C4作物), 另一方面也暗示它们在某些功能上可能具有相似性。玉米、高粱、谷子的PEPC成员与拟南芥、马铃薯、花生、蓖麻、烟草PEPC成员相互嵌合在一起, 表明PEPC基因在单、双子叶植物分化以前就已经存在。另外发现一些同源基因对, 例如AtPEPC1和AtPEPC3、StPEPC和NtPEPC1、AhPEPC1和RcPEPC、SbPEPC和ZmPEPC2、SiPEPC1和SbPEPC1、SbPEPC3和ZmPEPC1, 这些相同或不同物种的PEPC基因聚在一起表明, 它们可能由共同祖先进化而来, 而且具有类似的生物功能。
2.4 SiPEPC家族基因苗期非生物逆境胁迫下表达分析
由图4可知, 谷子三叶一心期幼苗在4种胁迫处理下, 除SiPEPC4由于表达量低未检测到信号外, 其他5个SiPEPC基因在不同时间点表达水平变化趋势不完全相同。SiPEPC6在ABA胁迫处理下所有时间点表达量均下调, 其他基因表达量在4种胁迫处理过程中至少1个时间点有所上调。SiPEPC1在ABA-12 h、Cold-24 h、PEG-12 h、NaCl-12 h时其相对表达量达最高, 分别为对照的8.20、9.70、4.85、9.93倍; SiPEPC2在ABA-12 h、Cold-24 h、PEG-6 h、NaCl-6 h时其相对表达量达最高, 分别为对照的4.81、8.77、5.69、21.38倍; SiPEPC3在ABA-6 h、Cold-24 h、PEG-24 h、NaCl-6 h时其相对表达量达最高, 分别为对照的13.91、1.34、3.55、3.22倍; SiPEPC5在ABA-6 h、Cold-24 h、PEG-24 h、NaCl-12 h时其相对表达量达最高, 分别为对照的16.00、31.70、4.31、9.24倍; 而SiPEPC6在ABA-12 h、Cold-24 h、PEG-12 h、NaCl-12 h时其相对表达量达最高, 分别为对照的0.93、5.19、3.53、15.68倍。试验表明5个SiPEPC基因都参与了苗期谷子对逆境胁迫的响应。图4
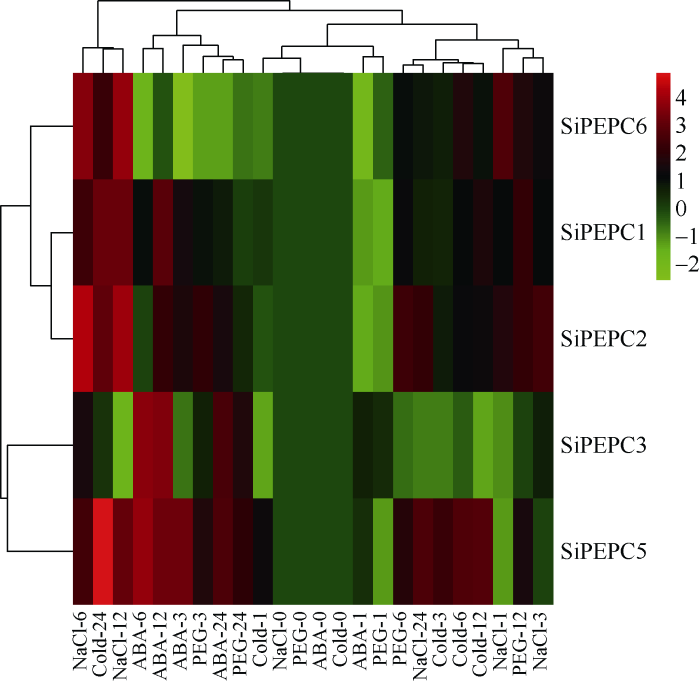 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4 SiPEPC基因苗期逆境表达谱
图中浅绿、深绿、黑色、浅红、深红5种颜色代表基因表达水平。绿色表示基因表达弱, 红色表示基因表达强。
Fig. 4Expression profile of SiPEPC gene under stresses at seedling stage
Light green, dark green, black, light red and dark red are used to represent gene expression levels. Green indicates weak gene expression; red indicates strong gene expression.
2.5 不同生育期干旱胁迫及不同光照强度下表达分析
为进一步了解PEPC基因在谷子不同生育期干旱胁迫下的表达情况, 本研究检测了它们在3个关键生育时期(拔节期、抽穗期和灌浆期)干旱胁迫及不同光照强度下的表达情况。由图5可知, SiPEPC1、SiPEPC2在正常抽穗期、灌浆期表达量与拔节期相比均有明显提升, 而SiPEPC3、SiPEPC5、SiPEPC6在抽穗期表达量下降, 在灌浆期表达量较拔节期有明显提升。干旱胁迫下, SiPEPC1、SiPEPC2、SiPEPC3在抽穗期、灌浆期表达量与拔节期相比均也有明显提升, SiPEPC5在灌浆期、抽穗期表达量与拔节期相比呈现先升后降趋势, 而SiPEPC6呈现先降后升趋势。表明大部分SiPEPC基因在正常生长条件下其表达随谷子生长发育进程呈现增强趋势, 在不同生育期干旱胁迫下其表达量趋势都有明显增加, 揭示这5个SiPEPC基因在关键生育时期都参与了对干旱胁迫的响应。图5
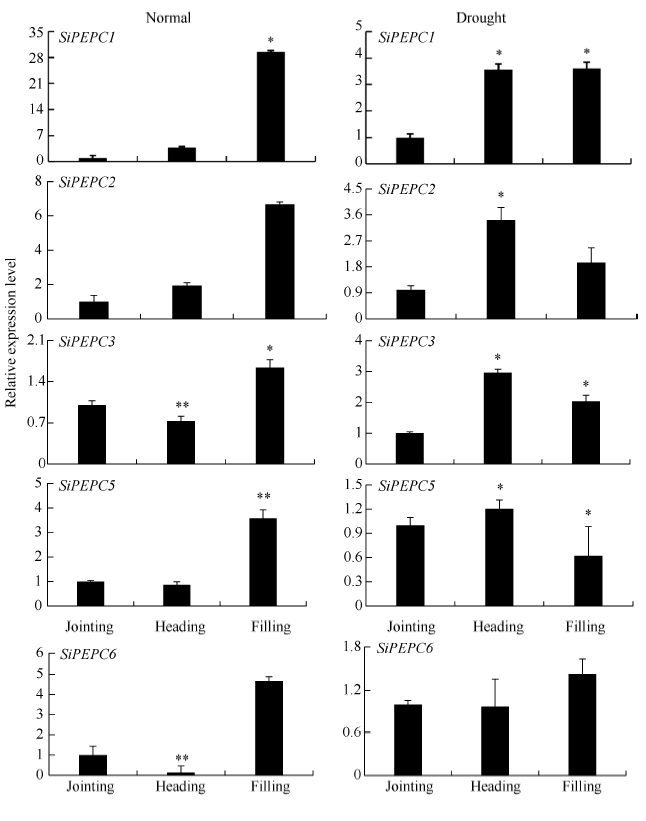 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5正常和干旱条件下SiPEPC基因在不同发育阶段的相对表达量
*表示在0.05水平上显著; **表示在0.01水平上显著。
Fig. 5Relative expression of SiPEPCs during different growth stages under normal and drought conditions
* Significantly different at P<0.05; ** Significantly different at P<0.01.
由图6可知, 所有SiPEPC基因热图颜色从J-1期(拔节期对照)偏黑红色向在J-2期(拔节期干旱)的鲜红色转变, 即所有基因在J-2期表达量与J-1期相比均有所提高, 也就是说拔节期干旱诱导了所有SiPEPC基因的表达。但在J-3期(拔节期中等光照)条件下所有基因表达量全部下降, 而在J-4期(拔节期弱光照)所有基因表达量又全部上升。试验结果说明拔节期光照强度影响SiPEPC基因表达, 弱光条件能诱导SiPEPC基因在拔节期的表达。与拔节期相似, 在H-2期(抽穗期干旱)所有基因表达量较对照H-1期(抽穗期对照)均有明显提高, 但在H-3 (抽穗期中等光照)和H-4 (抽穗期弱光照)条件下所有基因表达量都是下降的。说明抽穗期干旱诱导了所有SiPEPC基因的表达, 抽穗期中等、弱光照抑制SiPEPC基因的表达。在灌浆期的各个处理下(F-1、F-2、F-3、F-4), 除了SiPEPC3在F-2期(灌浆期)干旱表达量提升外, 其他所有基因在各处理下表达量均呈下降趋势。说明灌浆期大部分SiPEPC基因在干旱、中等和弱光照处理下表达量下降, 揭示SiPEPC基因在灌浆期可能对干旱、中等光照以及弱光照响应较弱。
图6
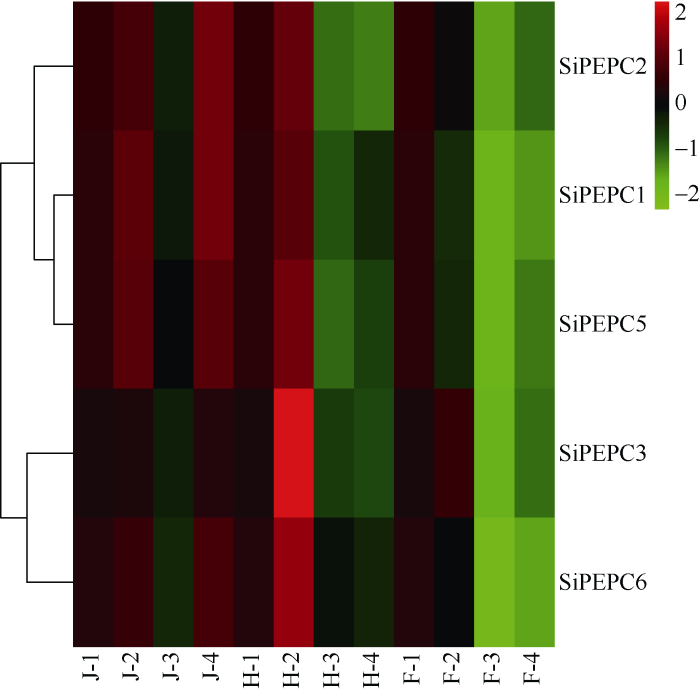 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6光照和干旱条件下SiPEPC 的相对表达量
图中浅绿、深绿、黑色、浅红、深红五色代表基因表达水平。绿色表示基因表达弱, 红色表示基因表达强。J-1: 拔节期对照; J-2: 拔节期干旱; J-3: 拔节期中等光照; J-4: 拔节期弱光照; H-1: 抽穗期对照; H-2: 抽穗期干旱; H-3: 抽穗期中等光照; H-4: 抽穗期弱光照; F-1: 灌浆期对照; F-2: 灌浆期干旱; F-3: 灌浆期中等光照; F-4: 灌浆期弱光照。
Fig. 6Relative expression of SiPEPC under light and drought conditions
Light green, dark green, black, light red, and dark red are used to represent gene expression levels. Green indicates weak gene expression; red indicates strong gene expression. J-1: control at jointing stage; J-2: drought at jointing stage; J-3: medium light intensity at jointing stage; J-4: weak light intensity at jointing stage; H-1: control at heading stage; H-2: drought at heading stage; H-3: medium light intensity at heading stage; H-4: weak light intensity at heading stage; F-1: drought at filling stage; F-2: drought at filling stage; F-3: medium light intensity at filling stage; F-4: weak light intensity at filling stage.
3 讨论
本研究在谷子基因组中检索得到6个SiPEPC基因, 序列比对显示, 谷子PEPC基因序列相似性很高, 揭示它们在进化上非常保守。系统进化树(图3)表明, 在单、双子叶植物分离以前PEPC基因就已经存在。进化树中谷子成员多与高粱、玉米成员紧密嵌合在一起, 而与拟南芥、马铃薯、花生、蓖麻、烟草成员相对较远, 揭示了谷子与高粱、玉米亲缘关系相对较近, 而与而与拟南芥、马铃薯、花生、蓖麻、烟草亲缘关系相对较远。值得一提的是, SiPEPC4基因由于在进化过程中存在一些片段的插入和缺失, 而且在基因组成上含有20个内含子, 远比其他SiPEPC基因内含子数目多, 因此推测SiPEPC4在功能上可能与其他SiPEPC基因有所不同。随后的表达分析也证明了这一点, 在所有表达谱分析中都检测不到SiPEPC4基因的表达, 说明SiPEPC4与其他SiPEPC基因在功能上有明显不同, 可能参与了谷子生长与发育中的其他信号途径。另外在进化树中发现一些同源基因对, 揭示这些同源基因对可能由共同祖先进化而来, 另一方面也暗示它们在某些信号通路中可能具有相似的功能。随后本研究对谷子6个SiPEPC基因生物信息学特征进行了系统的预测和分析, 期望能够发现对于功能研究有帮助信息。预测结果显示, SiPEPC成员很多性状和参数非常接近类似, 比如亚细胞定位分析SiPEPC成员都主要被定位在细胞质、细胞核和线粒体, 这些位置都是植物光合作用的关键位置, 揭示了SiPEPC基因可能在植物光合作用信号途径中起比较重要作用。这些结论不仅验证了它们同属PEPC基因家族, 而且预示不同SiPEPC基因可能共同参与或调控某些信号途径。顺式元件分析表明, 在SiPEPC启动子区域含有ABA、低温、干旱和防御应激反应等顺式元件。一般来讲如果基因启动子区域存在某种顺式元件则暗示该基因很可能参与相应的信号途径[33]。研究表明在非生物逆境, 比如干旱、盐、低温、高温以及伤害等胁迫下会导致植物细胞内的ABA水平升高, 而且众所周知, ABA是在非生物逆境应答中起重要作用的激素[34,35,36]。因此我们推测谷子SiPEPC家族基因很可能参与植物对非生物逆境胁迫的响应。
Real-time PCR检测结果表明, 5个谷子SiPEPC 基因在4种处理下其表达量有显著变化, 除了几乎所有基因表达量在4种胁迫处理过程中至少1个时间点均有所上调。试验结果表明, 谷子SiPEPC家族基因广泛参与了植物苗期非生物逆境胁迫应答。本研究结果同已有关于PEPC在逆境应答方面研究结果是相一致的[19,20,21]。为了进一步了解SiPEPC在不同关键生育期干旱胁迫下的表达情况, 本研究检测了SiPEPC成员在关键生育期(拔节期、抽穗期和灌浆期)干旱胁迫及不同光照强度(中等光照强度Ⅰ和弱光照强度Ⅱ)下的表达情况。如图5所示, 试验结果表明SiPEPC基因成员不同处理下表达情况各有特点。例如在正常条件下SiPEPC1、SiPEPC2在抽穗期、灌浆期表达量较拔节期均有明显提升, 而SiPEPC3、SiPEPC5、SiPEPC6在抽穗期表达下降, 在灌浆期表达明显提升。SiPEPC1、SiPEPC2、SiPEPC3在干旱胁迫下抽穗期、灌浆期表达较拔节期也有明显提升, SiPEPC5在灌浆期、抽穗期表达量呈现先升后降趋势, 而SiPEPC6呈现先降后升趋势。结果揭示SiPEPC基因在正常生长条件下随着谷子生长发育进程其表达趋势增强, 在不同生育期干旱胁迫下其表达量趋势都明显增加, 表明5个SiPEPC基因在不同生育期都参与了对干旱胁迫下的响应。不同生育期干旱、不同光照处理下表达分析(图6)表明, 所有SiPEPC基因在J-2期(拔节期干旱)表达量提高, 但在J-3期(拔节期中等光照)全部下降, 而在J-4期(拔节期弱光照)又全部上升。在H-2期(抽穗期干旱)所有基因表达量均有明显提高, 但在H-3 (抽穗期中等光照) H-4 (抽穗期弱光照)条件下都是下降的。在灌浆期的各个处理下(F-1、F-2、F-3、F-4), 只有SiPEPC3在F-2期(灌浆期)干旱表达量提升, 其他所有基因在各处理下表达量均呈下降趋势。试验结果说明拔节期干旱和弱光条件能诱导SiPEPC基因的表达, 抽穗期干旱诱导了所有SiPEPC基因的表达, 灌浆期大部分SiPEPC基因在干旱、中等和弱光照处理下表达量下降, SiPEPC基因参与了拔节期和抽穗期的干旱胁迫响应, 不同光照强度影响SiPEPC基因在不同生育期的表达, 弱光可诱导SiPEPC基因在拔节期的表达。光强对PEPC有调节作用, 穗期玉米遮阴后其叶片的PEPC活性显著降低, 且随光照强度的减弱而加剧, 穗期遮阴结束后PEPC活性能恢复到对照水平[37]。本研究结果只表明弱光可诱导SiPEPC基因在拔节期的表达, 未出现随光照强度的减弱而诱导加剧情况, 而在谷子抽穗期和灌浆期大部分SiPEPC基因没有检测到弱光能诱导PEPC表达的现象。这可能是谷子与玉米物种差别或是PEPC基因在不同生育期功能差别造成, 需要我们下一步详尽的试验验证, 但是这些结果进一步验证了顺式元件分析结果, 证明了SiPEPC基因在植物逆境应答中起一定作用。顺式元件分析还显示在SiPEPC基因启动子区域存在茉莉酸甲酯、水杨酸、赤霉素顺式原件。研究表明病原体感染通常导致细胞中茉莉酸甲酯、水杨酸、乙烯等激素水平的增加[38], 因此推断SiPEPC基因可能在生物应激反应中起一些作用。此外还发现了缺氧特异性诱导(GC-motif)、分生组织表达(CAT-box)、厌氧诱导必需(ARE)、玉米醇溶蛋白代谢调控(O2-site)、种子特异性调控(RY-element)、昼夜节律(circadian)等核心元件, 这些顺式元件的存在暗示PEPC可能参与相应的生理生化过程。需要注意的是在启动子区域还发现了大量的光应答元件, 众所周知谷子是光温敏感性作物, 而且PEPC是光合作用中的关键酶, 预示SiPEPC可能参与调控谷子的光温应答调控并在其中起重要作用。
本文报道的谷子SiPEPC基因也为进一步阐明SiPEPC在谷子逆境应答中的功能、机制提供了试验依据。尽管这些这些结论与推测需进一步严谨详尽的试验证明, 但是仍然为我们理解C4植物的关键酶PEPC在非生物逆境下的表达特征及光强对植物PEPC基因表达特性影响提供有益线索, 对深入了解C4植物光合特点对提高C3植物的光合效率和对C3植物的改造将具有一定意义。
4 结论
从谷子全基因组中鉴定出6个PEPC基因成员, 集中分布在谷子的1号、2号、4号、5号染色体上。所有蛋白序列高度保守, 都具有PEPC基因特征保守功能域PEPcase。谷子PEPC家族基因成员主要存在于细胞质、细胞核和线粒体。SiPEPC家族基因广泛参与了谷子苗期非生物逆境胁迫应答。SiPEPC成员参与了谷子在拔节、抽穗、灌浆期的干旱应答, 而光照强度会严重影响SiPEPC基因在不同生育期的表达。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/415451aURLPMID:11807559 [本文引用: 1]

Most plants are known as C3 plants because the first product of photosynthetic CO2 fixation is a three-carbon compound. C4 plants, which use an alternative pathway in which the first product is a four-carbon compound, have evolved independently many times and are found in at least 18 families. In addition to differences in their biochemistry, photosynthetic organs of C4 plants show alterations in their anatomy and ultrastructure. Little is known about whether the biochemical or anatomical characteristics of C4 photosynthesis evolved first. Here we report that tobacco, a typical C3 plant, shows characteristics of C4 photosynthesis in cells of stems and petioles that surround the xylem and phloem, and that these cells are supplied with carbon for photosynthesis from the vascular system and not from stomata. These photosynthetic cells possess high activities of enzymes characteristic of C4 photosynthesis, which allow the decarboxylation of four-carbon organic acids from the xylem and phloem, thus releasing CO2 for photosynthesis. These biochemical characteristics of C4 photosynthesis in cells around the vascular bundles of stems of C3 plants might explain why C4 photosynthesis has evolved independently many times.
DOI:10.1104/pp.106.085829URLPMID:16920871 [本文引用: 1]

Spatial and temporal regulation of phosphoenolpyruvate carboxylase (PEPC) is critical to the function of C(4) photosynthesis. The photosynthetic isoform of PEPC in the cytosol of mesophyll cells in Kranz-type C(4) photosynthesis has distinctive kinetic and regulatory properties. Some species in the Chenopodiaceae family perform C(4) photosynthesis without Kranz anatomy by spatial separation of initial fixation of atmospheric CO(2) via PEPC from C(4) acid decarboxylation and CO(2) donation to Rubisco within individual chlorenchyma cells. We studied molecular and functional features of PEPC in two single-cell functioning C(4) species (Bienertia sinuspersici, Suaeda aralocaspica) as compared to Kranz type (Haloxylon persicum, Salsola richteri, Suaeda eltonica) and C(3) (Suaeda linifolia) chenopods. It was found that PEPC from both types of C(4) chenopods displays higher specific activity than that of the C(3) species and shows kinetic and regulatory characteristics similar to those of C(4) species in other families in that they are subject to light/dark regulation by phosphorylation and display differential malate sensitivity. Also, the deduced amino acid sequence from leaf cDNA indicates that the single-cell functioning C(4) species possesses a Kranz-type C(4) isoform with a Ser in the amino terminal. A phylogeny of PEPC shows that isoforms in the two single-cell functioning C(4) species are in a clade with the C(3) and Kranz C(4) Suaeda spp. with high sequence homology. Overall, this study indicates that B. sinuspersici and S. aralocaspica have a C(4)-type PEPC similar to that in Kranz C(4) plants, which likely is required for effective function of C(4) photosynthesis.
DOI:10.1007/bf00021542URLPMID:8251640 [本文引用: 1]

A cDNA coding for phosphoenolpyruvate carboxylase (PEPC) was isolated from a cDNA library from Solanum tuberosum and the sequence of the cDNA was determined. It was inserted into a bacterial expression vector and a PEPC- Escherichia coli mutant could be complemented by the cDNA construct. A functional fusion protein could be synthesized in E. coli. The properties of this PEPC protein clearly resembled those of typical C3 plant enzymes.
DOI:10.1186/1752-0509-5-171URLPMID:22024416 [本文引用: 1]

C(4) plants such as corn and sugarcane assimilate atmospheric CO(2) into biomass by means of the C(4) carbon fixation pathway. We asked how PEP formation rate, a key step in the carbon fixation pathway, might work at a precise rate, regulated by light, despite fluctuations in substrate and enzyme levels constituting and regulating this process.
URL [本文引用: 1]

植物磷酸烯醇式丙酮酸羧化酶 (Phosphoenolpyruvate carboxylase,PEPC,EC 4.1.1.31) 是广泛存在的一种细胞质酶,催化磷酸烯醇式丙酮酸 (PEP) 和 HCO3-生成草酰乙酸 (OAA),后者可转化生成三羧酸循环的多种中间产物。PEPC在植物细胞中参与植物的光合碳同化等重要代谢途径,并且在不同组织中具有多种生理功能。PEPC同时也参与调控植物种子的营养物质合成与代谢过程,控制糖类物质流向脂肪酸合成或蛋白质合成途径。以下介绍了植物PEPC的种类、蛋白质结构特点及其在植物组织中的调控方式,并重点论述了PEPC在生物基因工程中的应用方面的进展,随着对其功能机制和应用研究的深入,将有助于植物PEPC在高产优质农作物育种、能源植物和工业微生物等的开发利用等方面得到更好的发展与应用。
URL [本文引用: 1]

植物磷酸烯醇式丙酮酸羧化酶 (Phosphoenolpyruvate carboxylase,PEPC,EC 4.1.1.31) 是广泛存在的一种细胞质酶,催化磷酸烯醇式丙酮酸 (PEP) 和 HCO3-生成草酰乙酸 (OAA),后者可转化生成三羧酸循环的多种中间产物。PEPC在植物细胞中参与植物的光合碳同化等重要代谢途径,并且在不同组织中具有多种生理功能。PEPC同时也参与调控植物种子的营养物质合成与代谢过程,控制糖类物质流向脂肪酸合成或蛋白质合成途径。以下介绍了植物PEPC的种类、蛋白质结构特点及其在植物组织中的调控方式,并重点论述了PEPC在生物基因工程中的应用方面的进展,随着对其功能机制和应用研究的深入,将有助于植物PEPC在高产优质农作物育种、能源植物和工业微生物等的开发利用等方面得到更好的发展与应用。
DOI:10.1016/j.plantsci.2006.09.008URL [本文引用: 1]
DOI:10.1093/pcp/pcu189URLPMID:25505033 [本文引用: 1]

Phosphoenolpyruvate carboxylase (PEPC) is a key enzyme of primary metabolism in bacteria, algae and vascular plants, and it undergoes allosteric regulation by various metabolic effectors. Rice (Oryza sativa) has five plant-type PEPCs, four cytosolic and one chloroplastic. We investigated their kinetic properties using recombinant proteins and found that, like most plant-type PEPCs, rice cytosolic isozymes were activated by glucose 6-phosphate and by alkaline pH. In contrast, no such activation was observed for the chloroplastic isozyme, Osppc4. In addition, Osppc4 showed low affinity for the substrate phosphoenolpyruvate (PEP) and very low sensitivities to allosteric inhibitors aspartate and glutamate. By comparing the isozyme amino acid sequences and three-dimensional structures simulated on the basis of the reported crystal structures, we identified two regions where Osppc4 has unique features that can be expected to affect its kinetic properties. One is the N-terminal extension; replacement of the extension of Osppc2a (cytosolic) with that from Osppc4 reduced the aspartate and glutamate sensitivities to about one-tenth of the wild-type values but left the PEP affinity unaffected. The other is the N-terminal loop, in which a conserved lysine at the N-terminal end is replaced with a glutamate-alanine pair in Osppc4. Replacement of the lysine of Osppc2a with glutamate-alanine lowered the PEP affinity to a quarter of the wild-type level (down to the Osppc4 level), without affecting inhibitor sensitivity. Both the N-terminal extension and the N-terminal loop are specific to plant-type PEPCs, suggesting that plant-type isozymes acquired these regions so that their activity could be regulated properly at the sites where they function.
DOI:10.1104/pp.102.019653URLPMID:12805623 [本文引用: 1]

Phosphoenolpyruvate carboxylase (PEPC) is distributed in plants and bacteria but is not found in fungi and animal cells. Important motifs for enzyme activity and structure are conserved in plant and bacterial PEPCs, with the exception of a phosphorylation domain present at the N terminus of all plant PEPCs reported so far, which is absent in the bacterial enzymes. Here, we describe a gene from Arabidopsis, stated as Atppc4, encoding a PEPC, which shows more similarity to Escherichia coli than to plant PEPCs. Interestingly, this enzyme lacks the phosphorylation domain, hence indicating that it is a bacterial-type PEPC. Three additional PEPC genes are present in Arabidopsis, stated as Atppc1, Atppc2, and Atppc3, encoding typical plant-type enzymes. As most plant PEPC genes, Atppc1, Atppc2, and Atppc3 are formed by 10 exons interrupted by nine introns. In contrast, Atppc4 gene has an unusual structure formed by 20 exons. A bacterial-type PEPC gene was also identified in rice (Oryza sativa), stated as Osppc-b, therefore showing the presence of this type of PEPC in monocots. The phylogenetic analysis suggests that both plant-type and bacterial-type PEPCs diverged early during the evolution of plants from a common ancestor, probably the PEPC from gamma-proteobacteria. The diversity of plant-type PEPCs in C3, C4, and Crassulacean acid metabolism plants is indicative of the evolutionary success of the regulation by phosphorylation of this enzyme. Although at a low level, the bacterial-type PEPC genes are expressed in Arabidopsis and rice.
DOI:10.1093/oxfordjournals.pcp.a029446URLPMID:9787461 [本文引用: 1]

A full-length cDNA for maize root-form phosphoenolpyruvate carboxylase (PEPC) was isolated. In the coding region, the root-form PEPC showed 76 and 77% identity with the C4- and C3-form PEPCs of maize, respectively, at the nucleotide level. At the amino acid level, the root-form was 81 and 85% identical to the C4- and C3-form PEPCs, respectively. The entire coding region was inserted into a pET32a expression vector so that it was expressed under the control of T7 promoter. The purified recombinant root-form PEPC had a Vmax value of about 28 mumol min-1 (mg protein)-1 at pH 8.0. The K(m) values of root-form PEPC for PEP and Mg2+ were one-tenth or less of those of C4-form PEPC when assayed at either pH 7.3 or 8.0, while the value for HCO3- was about one-half of that of C4-form PEPC at pH 8.0. Glucose 6-phosphate and glycine had little effect on the root-form PEPC at pH 7.3; they caused two-fold activation of the C4-form PEPC. The Ki (L-malate) values at pH 7.3 were 0.12 and 0.43 mM for the root- and C4-form PEPCs, respectively. Comparison of hydropathy profiles among the maize PEPC isoforms suggested that several stretches of amino acid sequences may contribute in some way to their characteristic kinetic properties. The root-form PEPC was phosphorylated by both mammalian cAMP-dependent protein kinase and maize leaf protein kinase, and the phosphorylated enzyme was less sensitive to L-malate.
DOI:10.1007/s00122-003-1268-2URLPMID:12759729 [本文引用: 1]

Phosphoenolpyruvate carboxylases (PEPCs) are encoded by a small multigenic family. In order to characterise this gene family in sugarcane, seven DNA fragments displaying a high homology with grass PEPC genes were isolated using polymerase chain reaction-based cloning. A phylogenetic study revealed the existence of four main PEPC gene lineages in grasses and particularly in sugarcane. Moreover, this analysis suggests that grass C4 PEPC has likely derived from a root pre-existing isoform in an ancestral species. Using the Northern-dot-blot method, we studied the expression of the four PEPC gene classes in sugarcane cv. R570. We confirmed that transcript accumulation of the C4 PEPC gene (ppc-C4) mainly occurs in the green leaves and is light-induced. We also showed that another member of this gene family (ppc-aR) is more highly transcribed in the roots. The constitutive expression for a previously characterised gene (ppc-aL2) was confirmed. Lastly, the transcript accumulation of the fourth PEPC gene class (ppc-aL1) was not revealed. Length polymorphism in non-coding regions for three PEPC gene lineages enabled us to develop sequence-tagged site PEPC markers in sugarcane. We analysed the segregation of PEPC fragments in self-pollinated progenies of cv. R570 and found co-segregating fragments for two PEPC gene lineages. This supports the hypothesis that diversification of the PEPC genes involved duplications, probably in tandem.
DOI:10.1104/pp.104.042762URLPMID:15299132 [本文引用: 1]

Phosphorylation of phosphoenolpyruvate carboxylase (PEPc; EC 4.1.1.31) plays an important role in the control of central metabolism of higher plants. This phosphorylation is controlled largely at the level of expression of PEPc kinase (PPCK) genes. We have analyzed the expression of both PPCK genes and the PEPC genes that encode PEPc in soybean (Glycine max). Soybean contains at least four PPCK genes. We report the genomic and cDNA sequences of these genes and demonstrate the function of the gene products by in vitro expression and enzyme assays. For two of these genes, GmPPCK2 and GmPPCK3, transcript abundance is highest in nodules and is markedly influenced by supply of photosynthate from the shoots. One gene, GmPPCK4, is under robust circadian control in leaves but not in roots. Its transcript abundance peaks in the latter stages of subjective day, and its promoter contains a sequence very similar to the evening element found in Arabidopsis genes expressed at this time. We report the expression patterns of five PEPC genes, including one encoding a bacterial-type PEPc lacking the phosphorylation site of the plant-type PEPcs. The PEPc expression patterns do not match those of any of the PPCK genes, arguing against the existence of specific PEPc-PPCK expression partners. The PEPC and PPCK gene families in soybean are significantly more complex than previously understood.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.plantsci.2007.02.016URL [本文引用: 1]
DOI:10.3724/SP.J.1006.2012.00285URL [本文引用: 1]

以过表达磷酸烯醇式丙酮酸羧化酶(PEPC)的水稻为材料,研究了不同程度干旱胁迫下开花期剑叶光合作用的光响应过程、叶绿素荧光参数、色素含量和活性氧代谢。结果表明,在干旱特别是重度干旱胁迫下,野生型水稻在强光下净光合速率迅速下降,而转Zmppc基因水稻没有明显的下降现象; 而且表示光化学活性的叶绿素荧光参数Fv/Fm、ΦPSⅡ和qP下降程度低,说明PEPC增强了干旱胁迫下水稻抵御强光胁迫的能力。这可能是因为干旱胁迫下转Zmppc基因水稻玉米黄质含量高,光系统对过剩光能的耗散能力强,能够保护光系统免受过剩光能的伤害,从而减小O2?产生速率; 同时干旱胁迫下转PEPC基因水稻抗氧化酶SOD、POD和CAT活性高,能有效清除活性氧,减轻膜质过氧化。
DOI:10.3724/SP.J.1006.2012.00285URL [本文引用: 1]

以过表达磷酸烯醇式丙酮酸羧化酶(PEPC)的水稻为材料,研究了不同程度干旱胁迫下开花期剑叶光合作用的光响应过程、叶绿素荧光参数、色素含量和活性氧代谢。结果表明,在干旱特别是重度干旱胁迫下,野生型水稻在强光下净光合速率迅速下降,而转Zmppc基因水稻没有明显的下降现象; 而且表示光化学活性的叶绿素荧光参数Fv/Fm、ΦPSⅡ和qP下降程度低,说明PEPC增强了干旱胁迫下水稻抵御强光胁迫的能力。这可能是因为干旱胁迫下转Zmppc基因水稻玉米黄质含量高,光系统对过剩光能的耗散能力强,能够保护光系统免受过剩光能的伤害,从而减小O2?产生速率; 同时干旱胁迫下转PEPC基因水稻抗氧化酶SOD、POD和CAT活性高,能有效清除活性氧,减轻膜质过氧化。
URL [本文引用: 1]

以转PEPC,PPDK,NADP-ME,PEPC+PPDK基因水稻及原种为材料,比较C4途径有关光合酶活性、叶绿素荧光参数、CO2交换等生理指标,重点研究了转PEPC基因水稻的生理特性, 结果如下:(1)转PEPC基因水稻PEPC活性比原种高20倍,光饱和光合速率比原种高55%,羧化效率提高50%,CO2补偿点降低27%;(2)在高光强(3 h)或光氧化剂甲基紫精(MV)处理后, 与原种相比,转PEPC基因水稻的PSⅡ光化学效率(Fv /Fm)、光化学猝灭(qP)下降较少,证明其耐光抑制和耐光氧化能力增强;(3)在高光强条件下,转PEPC基因水稻中RuBPCase活性变化不明显,但碳酸酐酶(CA)诱导活性增加1.8倍.这些结果为揭示高光合效率机理和高光合效率育种提供了有益的启示.
URL [本文引用: 1]

以转PEPC,PPDK,NADP-ME,PEPC+PPDK基因水稻及原种为材料,比较C4途径有关光合酶活性、叶绿素荧光参数、CO2交换等生理指标,重点研究了转PEPC基因水稻的生理特性, 结果如下:(1)转PEPC基因水稻PEPC活性比原种高20倍,光饱和光合速率比原种高55%,羧化效率提高50%,CO2补偿点降低27%;(2)在高光强(3 h)或光氧化剂甲基紫精(MV)处理后, 与原种相比,转PEPC基因水稻的PSⅡ光化学效率(Fv /Fm)、光化学猝灭(qP)下降较少,证明其耐光抑制和耐光氧化能力增强;(3)在高光强条件下,转PEPC基因水稻中RuBPCase活性变化不明显,但碳酸酐酶(CA)诱导活性增加1.8倍.这些结果为揭示高光合效率机理和高光合效率育种提供了有益的启示.
URL [本文引用: 1]

通过C4转基因技术改善C3作物光合作用, 以期提高作物产量是国内外研究的热点之一。然而, 目前关于转磷酸烯醇式丙酮酸羧化酶(PEPC)基因对水稻的光合作用、产量和抗旱性的影响及其调节机理仍不很清楚。本研究以T4代转ppc基因水稻为材料, 进行产量和苗期抗旱性研究。结果表明, 在旱作栽培条件下, 两个转ppc基因株系(T1, T2)单株产量分别比未转基因的对照(WT)增产28%和42%, 分蘖增加27%和40%; T1和T2可维持较高的光合速率、气孔导度和水分利用效率; 干旱胁迫使超氧化物歧化酶(SOD)活性和丙二醛(MDA)含量增加, T1和T2 SOD活性增幅(+25%)都显著高于WT(+9%), 而MDA含量增幅显著低于WT, 表明转ppc基因水稻具有较强的抗氧化能力; T1和T2比WT具有较高的脯氨酸含量和渗透势下降幅度, 说明转PEPC基因水稻的渗透调节能力高于对照。PEG-6000处理下, 得到的结果相似。
URL [本文引用: 1]

通过C4转基因技术改善C3作物光合作用, 以期提高作物产量是国内外研究的热点之一。然而, 目前关于转磷酸烯醇式丙酮酸羧化酶(PEPC)基因对水稻的光合作用、产量和抗旱性的影响及其调节机理仍不很清楚。本研究以T4代转ppc基因水稻为材料, 进行产量和苗期抗旱性研究。结果表明, 在旱作栽培条件下, 两个转ppc基因株系(T1, T2)单株产量分别比未转基因的对照(WT)增产28%和42%, 分蘖增加27%和40%; T1和T2可维持较高的光合速率、气孔导度和水分利用效率; 干旱胁迫使超氧化物歧化酶(SOD)活性和丙二醛(MDA)含量增加, T1和T2 SOD活性增幅(+25%)都显著高于WT(+9%), 而MDA含量增幅显著低于WT, 表明转ppc基因水稻具有较强的抗氧化能力; T1和T2比WT具有较高的脯氨酸含量和渗透势下降幅度, 说明转PEPC基因水稻的渗透调节能力高于对照。PEG-6000处理下, 得到的结果相似。
DOI:10.1016/S0300-9084(02)00024-XURL [本文引用: 1]

Abstract
Water availability is one of the major limiting factors for plant growth. Maize is particularly sensitive to water stress at reproductive stages with a strong impairment of photosynthesis and grain filling. Here, we describe the use of genetic transformation first to assess the role of a candidate gene Asr1—a putative transcription factor—as an explanation for genetically linked drought tolerance Quantitative Trait Loci (QTLs), and second to modify CO2 fixation rates in leaves through changes of C4 phosphoenolpyruvate carboxylase (C4-PEPC) activity. Transgenic Asr1 over-expressing lines show an increase in foliar senescence under drought conditions. The highest C4-PEPC overexpressing line exhibited an increase (+30%) in intrinsic water use efficiency (WUE) accompanied by a dry weight increase (+20%) under moderate drought conditions. Opposite effects were observed for transgenic plants under-expressing the corresponding proteins. The data presented here indicate the feasibility to increase the level of endogenous biochemical activities related to water economy and/or drought tolerance, and opens a way to develop maize varieties more tolerant to dry growing conditions.DOI:10.1007/s00425-002-0951-xURLPMID:12687366 [本文引用: 2]

Phosphoenolpyruvate carboxylase (PEPC, EC 4.1.1.31) plays an important role in CO(2) fixation in C4 and CAM plants. In C3 plants, PEPC is widely expressed in most organs; however, its function is not yet clearly established. With the aim of providing clues on the function of PEPC in C3 plants, we have analyzed its pattern of expression in wheat ( Triticum aestivum L.) seedlings. Roots showed almost double the level of PEPC activity of shoots. Further analysis of PEPC expression in roots by in situ localization techniques showed a high accumulation of PEPC transcripts and polypeptides in meristematic cells, whereas in the rest of the root PEPC localized preferentially to the vascular tissue. Treatment with NaCl and LiCl induced PEPC expression in roots. Similarly, other abiotic stresses affecting water status, such as drought or cold, induced PEPC expression. Induction was root-specific except for the cold treatment, which also induced PEPC in shoots, although to a lesser extent. In contrast, hypoxia, which does not affect water balance, did not promote any induction of PEPC expression. These results suggest an important role for this enzyme in the adaptation of plants to environmental changes.
DOI:10.1007/s00425-005-0144-5URL [本文引用: 2]

The phosphoenolpyruvate carboxylase (PEPC) gene family of Arabidopsis is composed of four genes. Based on sequence analysis it was deduced that Atppc1, Atppc2 and Atppc3 genes encode plant-type PEPCs, whereas Atppc4 encodes a PEPC without phosphorylation motif, but no data at the protein level have been reported. Here, we describe the analysis of the four Arabidopsis PEPC polypeptides, which were expressed in Escherichia coli. Immunological characterization with anti plant-type PEPC and an anti-AtPPC4 antibody, raised in this work, showed that the bacterial-type PEPC is unrelated with plant-type PEPCs. Western-blot analysis of different Arabidopsis organs probed with anti plant-type PEPC antibodies detected a double band, the one with low molecular weight corresponding to the three plant-type PEPCs. The high molecular weight subunit is not encoded by any of the Arabidopsis PEPC genes. No bands were detected with the anti-AtPPC4 antibody. PEPC genes show differential expression in Arabidopsis organs and in response to environmental stress. Atppc2 transcripts were found in all Arabidopsis organs suggesting that it is a housekeeping gene. In contrast, Atppc3 gene was expressed in roots and Atppc1 in roots and flowers, as Atppc4. Highest PEPC activity was found in roots, which showed expression of the four PEPC genes. Salt and drought exerted a differential induction of PEPC gene expression in roots, Atppc4 showing the highest induction in response to both stresses. These results show that PEPC is part of the adaptation of the plant to salt and drought and suggest that this is the function of the new bacterial-type PEPC.
DOI:10.1007/s00425-002-0893-3URLPMID:12569407 [本文引用: 2]

C(4) phosphoenolpyruvate carboxylase (PEPCase: EC 4.1.1.31) is subjected to in vivo regulatory phosphorylation by a light up-regulated, calcium-independent protein kinase. Salt stress greatly enhanced phosphoenolpyruvate carboxylase-kinase (PEPCase-k) activity in leaves of Sorghum. The increase in PEPCase-k anticipated the time course of proline accumulation thereby suggesting that water stress was not involved in the kinase response to salt. Moreover, osmotic stress seemed not to be the main factor implicated, as demonstrated by the lack of effect when water availability was restricted by mannitol. In contrast, LiCl (at a concentration of 10 mM in short-term treatment of both excised leaves and whole plants) mimicked the effects of 172 mM NaCl salt-acclimation, indicating that the rise in PEPCase-k activity resulted primarily from the ionic stress. Both NaCl and LiCl treatments increased the activity of a Ca(2+)-independent, 35 kDa kinase, as demonstrated by an in-gel phosphorylation experiment. Short-term treatment of excised leaves with NaCl or LiCl partially reproduces the effects of whole plant treatments. Finally, salinization also increased PEPCase-k activity and the phosphorylation state of PEPCase in darkened Sorghum leaves. This fact, together with increased malate production during the dark period, suggests a shift towards mixed C(4) and crassulacean acid metabolism types of photosynthesis in response to salt stress.
DOI:10.3724/SP.J.1006.2012.00800URL [本文引用: 1]

谷子是重要的饲草作物, 但其饲草品质性状研究滞后制约了谷子饲草品种培育和栽培。本研究对47个谷子品种在我国高寒农牧地区栽培的饲用干草品质性状进行了系统测定。结果表明, 供试材料粗蛋白含量的变异范围为5.42%~12.45%, 变异系数为14.25;粗脂肪含量的变异范围为0.64%~1.43%, 变异系数为15.84;粗灰分含量的变异范围为8.50%~15.60%, 变异系数为13.71;总磷含量的变异范围为0.10%~0.32%, 变异系数为20.00;总钙含量的变异范围为0.27%~0.67%, 变异系数为20.00。粗纤维、无氮浸出物和水分含量表现出较小的变异。主成分分析表明, 影响谷子饲草品质的主要性状是粗纤维、粗灰分、粗蛋白和无氮浸出物, 粗蛋白与粗纤维和无氮浸出物均为负相关。综合表明, Li05-569、饿死驴、系295和红根谷是品质性状综合表现优良的品种。
DOI:10.3724/SP.J.1006.2012.00800URL [本文引用: 1]

谷子是重要的饲草作物, 但其饲草品质性状研究滞后制约了谷子饲草品种培育和栽培。本研究对47个谷子品种在我国高寒农牧地区栽培的饲用干草品质性状进行了系统测定。结果表明, 供试材料粗蛋白含量的变异范围为5.42%~12.45%, 变异系数为14.25;粗脂肪含量的变异范围为0.64%~1.43%, 变异系数为15.84;粗灰分含量的变异范围为8.50%~15.60%, 变异系数为13.71;总磷含量的变异范围为0.10%~0.32%, 变异系数为20.00;总钙含量的变异范围为0.27%~0.67%, 变异系数为20.00。粗纤维、无氮浸出物和水分含量表现出较小的变异。主成分分析表明, 影响谷子饲草品质的主要性状是粗纤维、粗灰分、粗蛋白和无氮浸出物, 粗蛋白与粗纤维和无氮浸出物均为负相关。综合表明, Li05-569、饿死驴、系295和红根谷是品质性状综合表现优良的品种。
DOI:10.1007/s001220050709URL [本文引用: 1]
DOI:10.1007/s12033-008-9081-4URL [本文引用: 1]

Plant growth and productivity are affected by various abiotic stresses such as heat, drought, cold, salinity, etc. The mechanism of salt tolerance is one of the most important subjects in plant science as salt stress decreases worldwide agricultural production. In our present study we used cDNA-AFLP technique to compare gene expression profiles of a salt tolerant and a salt-sensitive cultivar of foxtail millet (Seteria italica) in response to salt stress to identify early responsive differentially expressed transcripts accumulated upon salt stress and validate the obtained result through quantitative real-time PCR (qRT-PCR). The expression profile was compared between a salt tolerant (Prasad) and susceptible variety (Lepakshi) of foxtail millet in both control condition (L0 and P0) and after 1h (L1 and P1) of salt stress. We identified 90 transcript-derived fragments (TDFs) that are differentially expressed, out of which 86 TDFs were classified on the basis of their either complete presence or absence (qualitative variants) and 4 on differential expression pattern levels (quantitative variants) in the two varieties. Finally, we identified 27 non-redundant differentially expressed cDNAs that are unique to salt tolerant variety which represent different groups of genes involved in metabolism, cellular transport, cell signaling, transcriptional regulation, mRNA splicing, seed development and storage, etc. The expression patterns of seven out of nine such genes showed a significant increase of differential expression in tolerant variety after 1h of salt stress in comparison to salt-sensitive variety as analyzed by qRT-PCR. The direct and indirect relationship of identified TDFs with salinity tolerance mechanism is discussed.
DOI:10.3724/SP.J.1006.2013.00360URL [本文引用: 1]

CBL/CIPK信号网络系统在植物对逆境应答过程中起重要作用。本文利用生物信息学方法从谷子基因组中鉴定出7个候选CBL基因,命名为SiCBL1~SiCBL7。分析表明谷子CBL基因在蛋白质序列和结构上非常保守,所有预测SiCBL基因都含有7~8个内含子,而且其外显子序列同源性很高并且大部分外显子含有相同的碱基数目。预测SiCBL基因均含有4个EF-Hand功能域而且相邻EF-Hand功能域之间的氨基酸数目非常保守。进化和聚类分析结果表明, CBL基因在陆生植物早期的进化过程中就已经存在,所有CBL基因被分为4个亚组。7个候选SiCBL基因中, 4个基因(SiCBL1、SiCBL2、SiCBL3和SiCBL5)受干旱胁迫诱导表达,3个(SiCBL1、SiCBL3和SiCBL7)受高盐胁迫诱导表达。SiCBL3基因在干旱胁迫下被强烈诱导可能意味着其在干旱应答中起重要作用。本文报道的谷子CBL基因丰富和完善了植物CBL成员, 为进一步研究CBL/CIPK网络系统在抗逆作物谷子逆境响应中的功能、机制奠定了基础。
DOI:10.3724/SP.J.1006.2013.00360URL [本文引用: 1]

CBL/CIPK信号网络系统在植物对逆境应答过程中起重要作用。本文利用生物信息学方法从谷子基因组中鉴定出7个候选CBL基因,命名为SiCBL1~SiCBL7。分析表明谷子CBL基因在蛋白质序列和结构上非常保守,所有预测SiCBL基因都含有7~8个内含子,而且其外显子序列同源性很高并且大部分外显子含有相同的碱基数目。预测SiCBL基因均含有4个EF-Hand功能域而且相邻EF-Hand功能域之间的氨基酸数目非常保守。进化和聚类分析结果表明, CBL基因在陆生植物早期的进化过程中就已经存在,所有CBL基因被分为4个亚组。7个候选SiCBL基因中, 4个基因(SiCBL1、SiCBL2、SiCBL3和SiCBL5)受干旱胁迫诱导表达,3个(SiCBL1、SiCBL3和SiCBL7)受高盐胁迫诱导表达。SiCBL3基因在干旱胁迫下被强烈诱导可能意味着其在干旱应答中起重要作用。本文报道的谷子CBL基因丰富和完善了植物CBL成员, 为进一步研究CBL/CIPK网络系统在抗逆作物谷子逆境响应中的功能、机制奠定了基础。
DOI:10.1038/nbt.2195URLPMID:22580950 [本文引用: 1]

Foxtail millet (Setaria italica), a member of the Poaceae grass family, is an important food and fodder crop in arid regions and has potential for use as a C(4) biofuel. It is a model system for other biofuel grasses, including switchgrass and pearl millet. We produced a draft genome (~423 Mb) anchored onto nine chromosomes and annotated 38,801 genes. Key chromosome reshuffling events were detected through collinearity identification between foxtail millet, rice and sorghum including two reshuffling events fusing rice chromosomes 7 and 9, 3 and 10 to foxtail millet chromosomes 2 and 9, respectively, that occurred after the divergence of foxtail millet and rice, and a single reshuffling event fusing rice chromosome 5 and 12 to foxtail millet chromosome 3 that occurred after the divergence of millet and sorghum. Rearrangements in the C(4) photosynthesis pathway were also identified.
DOI:10.1038/nbt.2196URLPMID:22580951 [本文引用: 1]

We generated a high-quality reference genome sequence for foxtail millet (Setaria italica). The ~400-Mb assembly covers ~80% of the genome and &gt;95% of the gene space. The assembly was anchored to a 992-locus genetic map and was annotated by comparison with &gt;1.3 million expressed sequence tag reads. We produced more than 580 million RNA-Seq reads to facilitate expression analyses. We also sequenced Setaria viridis, the ancestral wild relative of S. italica, and identified regions of differential single-nucleotide polymorphism density, distribution of transposable elements, small RNA content, chromosomal rearrangement and segregation distortion. The genus Setaria includes natural and cultivated species that demonstrate a wide capacity for adaptation. The genetic basis of this adaptation was investigated by comparing five sequenced grass genomes. We also used the diploid Setaria genome to evaluate the ongoing genome assembly of a related polyploid, switchgrass (Panicum virgatum).
DOI:10.1105/tpc.6.2.251URLPMID:8148648 [本文引用: 1]

Two genes, rd29A and rd29B, which are closely located on the Arabidopsis genome, are differentially induced under conditions of dehydration, low temperature, high salt, or treatment with exogenous abscisic acid (ABA). It appears that rd29A has at least two cis-acting elements, one involved in the ABA-associated response to dehydration and the other induced by changes in osmotic potential, and that rd29B contains at least one cis-acting element that is involved in ABA-responsive, slow induction. We analyzed the rd29A promoter in both transgenic Arabidopsis and tobacco and identified a novel cis-acting, dehydration-responsive element (DRE) containing 9 bp, TACCGACAT, that is involved in the first rapid response of rd29A to conditions of dehydration or high salt. DRE is also involved in the induction by low temperature but does not function in the ABA-responsive, slow expression of rd29A. Nuclear proteins that specifically bind to DRE were detected in Arabidopsis plants under either high-salt or normal conditions. Different cis-acting elements seem to function in the two-step induction of rd29A and in the slow induction of rd29B under conditions of dehydration, high salt, or low temperature.
DOI:10.1002/prot.21018URLPMID:16752418 [本文引用: 1]

Because the protein's function is usually related to its subcellular localization, the ability to predict subcellular localization directly from protein sequences will be useful for inferring protein functions. Recent years have seen a surging interest in the development of novel computational tools to predict subcellular localization. At present, these approaches, based on a wide range of algorithms, have achieved varying degrees of success for specific organisms and for certain localization categories. A number of authors have noticed that sequence similarity is useful in predicting subcellular localization. For example, Nair and Rost (Protein Sci 2002;11:2836-2847) have carried out extensive analysis of the relation between sequence similarity and identity in subcellular localization, and have found a close relationship between them above a certain similarity threshold. However, many existing benchmark data sets used for the prediction accuracy assessment contain highly homologous sequences-some data sets comprising sequences up to 80-90% sequence identity. Using these benchmark test data will surely lead to overestimation of the performance of the methods considered. Here, we develop an approach based on a two-level support vector machine (SVM) system: the first level comprises a number of SVM classifiers, each based on a specific type of feature vectors derived from sequences; the second level SVM classifier functions as the jury machine to generate the probability distribution of decisions for possible localizations. We compare our approach with a global sequence alignment approach and other existing approaches for two benchmark data sets-one comprising prokaryotic sequences and the other eukaryotic sequences. Furthermore, we carried out all-against-all sequence alignment for several data sets to investigate the relationship between sequence homology and subcellular localization. Our results, which are consistent with previous studies, indicate that the homology search approach performs well down to 30% sequence identity, although its performance deteriorates considerably for sequences sharing lower sequence identity. A data set of high homology levels will undoubtedly lead to biased assessment of the performances of the predictive approaches-especially those relying on homology search or sequence annotations. Our two-level classification system based on SVM does not rely on homology search; therefore, its performance remains relatively unaffected by sequence homology. When compared with other approaches, our approach performed significantly better. Furthermore, we also develop a practical hybrid method, which combines the two-level SVM classifier and the homology search method, as a general tool for the sequence annotation of subcellular localization.
DOI:10.1093/bioinformatics/btm404URLPMID:17846036 [本文引用: 1]

The Clustal W and Clustal X multiple sequence alignment programs have been completely rewritten in C++. This will facilitate the further development of the alignment algorithms in the future and has allowed proper porting of the programs to the latest versions of Linux, Macintosh and Windows operating systems.
DOI:10.1093/molbev/mst197URL [本文引用: 1]

We announce the release of an advanced version of the Molecular Evolutionary Genetics Analysis (MEGA) software, which currently contains facilities for building sequence alignments, inferring phylogenetic histories, and conducting molecular evolutionary analysis. In version 6.0, MEGA now enables the inference of timetrees, as it implements the RelTime method for estimating divergence times for all branching points in a phylogeny. A new Timetree Wizard in MEGA6 facilitates this timetree inference by providing a graphical user interface (GUI) to specify the phylogeny and calibration constraints step-by-step. This version also contains enhanced algorithms to search for the optimal trees under evolutionary criteria and implements a more advanced memory management that can double the size of sequence data sets to which MEGA can be applied. Both GUI and command-line versions of MEGA6 can be downloaded from www.megasoftware.net free of charge.
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
DOI:10.3897/zookeys.621.10115URLPMID:27833421 [本文引用: 1]

A new minute-size empidoid fly genus, Gondwanamyiagen. n. and two new species (Gondwanamyia chilensis Cumming & Saigusa, sp. n., Gondwanamyia zealandica Sinclair & Brooks, sp. n.) are described, illustrated, and their distributions mapped. The family and subfamily assignments remain uncertain, but features of the female terminalia potentially suggest Trichopezinae (Brachystomatidae).
DOI:10.1007/s00299-013-1418-1URL [本文引用: 1]

We review the recent progress on ABA signaling, especially ABA signaling for ABA-dependent gene expression, including the AREB/ABF regulon, SnRK2 protein kinase, 2C-type protein phosphatases and ABA receptors.
Drought negatively impacts plant growth and the productivity of crops. Drought causes osmotic stress to organisms, and the osmotic stress causes dehydration in plant cells. Abscisic acid (ABA) is produced under osmotic stress conditions, and it plays an important role in the stress response and tolerance of plants. ABA regulates many genes under osmotic stress conditions. It also regulates gene expression during seed development and germination. The ABA-responsive element (ABRE) is the major cis-element for ABA-responsive gene expression. ABRE-binding protein (AREB)/ABRE-binding factor (ABF) transcription factors (TFs) regulate ABRE-dependent gene expression. Other TFs are also involved in ABA-responsive gene expression. SNF1-related protein kinases 2 are the key regulators of ABA signaling including the AREB/ABF regulon. Recently, ABA receptors and group A 2C-type protein phosphatases were shown to govern the ABA signaling pathway. Moreover, recent studies have suggested that there are interactions between the major ABA signaling pathway and other signaling factors in stress-response and seed development. The control of the expression of ABA signaling factors may improve tolerance to environmental stresses.
DOI:10.1186/s12870-016-0771-yURLPMID:27079791 [本文引用: 1]

Being sessile organisms, plants are often exposed to a wide array of abiotic and biotic stresses. Abiotic stress conditions include drought, heat, cold and salinity, whereas biotic stress arises mainly from bacteria, fungi, viruses, nematodes and insects. To adapt to such adverse situations, plants have evolved well-developed mechanisms that help to perceive the stress signal and enable optimal growth response. Phytohormones play critical roles in helping the plants to adapt to adverse environmental conditions. The elaborate hormone signaling networks and their ability to crosstalk make them ideal candidates for mediating defense responses.
DOI:10.1016/j.fcr.2005.08.018URL [本文引用: 1]
URL [本文引用: 1]

在大田条件下研究不同时期和不同程度遮荫对夏玉米光合特性及其产量的影响。结果表明,遮荫对玉米叶片的光合特性有显著影响,导致玉米籽粒产量显著降低,农大108(ND108)苗期遮荫50%和90%的籽粒产量分别比对照降低16.9%和24.5%,穗期遮荫分别降低34.1%和55.3%,花粒期遮荫分别降低67.5%和79.4%。遮荫后玉米的叶面积指数、叶绿素含量和净光合速率等都显著降低。不同生育时期对遮荫的响应不同,苗期遮荫50%和90%处理的叶面积指数分别较对照降低53.6%和64.3%,穗期分别降低22.1%和23.0%,花粒期分别降低66.2%和79.4%。遮荫50%和90%处理叶片的光合速率苗期分别较对照降低28.6%和49.0%,穗期分别降低36.6%和59.6%,花粒期分别降低43.9%和64.7%。遮荫使玉米叶片的磷酸烯醇式丙酮酸羧化酶(PEPCase)和核酮糖二磷酸羧化酶(RuBPCase)活性显著降低,不同遮荫时期对其影响不同,花粒期遮荫影响最显著。遮荫时期对玉米光合特性的影响大于遮荫程度对其影响。
URL [本文引用: 1]

在大田条件下研究不同时期和不同程度遮荫对夏玉米光合特性及其产量的影响。结果表明,遮荫对玉米叶片的光合特性有显著影响,导致玉米籽粒产量显著降低,农大108(ND108)苗期遮荫50%和90%的籽粒产量分别比对照降低16.9%和24.5%,穗期遮荫分别降低34.1%和55.3%,花粒期遮荫分别降低67.5%和79.4%。遮荫后玉米的叶面积指数、叶绿素含量和净光合速率等都显著降低。不同生育时期对遮荫的响应不同,苗期遮荫50%和90%处理的叶面积指数分别较对照降低53.6%和64.3%,穗期分别降低22.1%和23.0%,花粒期分别降低66.2%和79.4%。遮荫50%和90%处理叶片的光合速率苗期分别较对照降低28.6%和49.0%,穗期分别降低36.6%和59.6%,花粒期分别降低43.9%和64.7%。遮荫使玉米叶片的磷酸烯醇式丙酮酸羧化酶(PEPCase)和核酮糖二磷酸羧化酶(RuBPCase)活性显著降低,不同遮荫时期对其影响不同,花粒期遮荫影响最显著。遮荫时期对玉米光合特性的影响大于遮荫程度对其影响。
DOI:10.1007/s11103-008-9435-0URL [本文引用: 1]

Plant hormones play important roles in regulating developmental processes and signaling networks involved in plant responses to a wide range of biotic and abiotic stresses. Significant progress has been made in identifying the key components and understanding the role of salicylic acid (SA), jasmonates (JA) and ethylene (ET) in plant responses to biotic stresses. Recent studies indicate that other hormones such as abscisic acid (ABA), auxin, gibberellic acid (GA), cytokinin (CK), brassinosteroids (BR) and peptide hormones are also implicated in plant defence signaling pathways but their role in plant defence is less well studied. Here, we review recent advances made in understanding the role of these hormones in modulating plant defence responses against various diseases and pests.
