

1. 安徽大学 物质科学与信息技术研究院,安徽 合肥 230601;
2. 安徽大学 化学与化工学院,安徽 合肥 230601;
3. 安徽大学 生命科学学院,安徽 合肥 230601
收稿日期:2020-08-11;接收日期:2020-12-11;网络出版时间:2021-01-12
基金项目:国家自然科学基金(Nos. 31972930,31570074),安徽高校协同创新项目(No. GXXT-2019-035) 资助
摘要:TetR家族转录调控因子(TetR family transcriptional regulators,TFRs) 广泛分布于细菌与古菌中,最早被发现的是可控制大肠杆菌四环素外排泵的TetR。TFRs拥有DNA结合和多样的配体感知能力,其作用机制复杂。配体小分子诱导TFRs产生构象变化,抑制或促进TFRs对其靶点的控制。目前已知的TFRs配体种类繁多,包括糖类、蛋白质、脂肪酸及其衍生物、金属离子等。配体的多样性使得TFRs调节范围很广,包括从基础碳代谢、氮代谢到群体感应、抗生素生物合成等一系列生理过程。文中主要从介导调控角度介绍TFRs配体的研究进展,并结合笔者实验室的研究工作,重点阐述TFRs配体在基础碳代谢、脂肪酸生物合成与降解等初级和次级代谢产物合成过程中的作用机制,及其在基因线路开发和抗生素合成基因的激活等应用方面的研究进展。
关键词:细菌古菌TetR家族转录因子配体反馈调控前馈控制
Ligands of TetR family transcriptional regulators: a review
Panpan Wu1,2, Bowen Li1, Ketao Chen1, Hang Wu1,3, Buchang Zhang1,2,3


1. Institutes of Physical Science and Information Technology, Anhui University, Hefei 230601, Anhui, China;
2. College of Chemistry and Chemical Engineering, Anhui University, Hefei 230601, Anhui, China;
3. School of Life Sciences, Anhui University, Hefei 230601, Anhui, China
Received: August 11, 2020; Accepted: December 11, 2020; Published: January 12, 2021
Supported by: National Natural Science Foundation of China (Nos. 31972930, 31570074), the University Synergy Innovation Program of Anhui Province, China (No. GXXT-2019-035)
Corresponding author: Buchang Zhang. Tel: +86-551-63861965; E-mail: zhbc@ahu.edu.cn.
Abstract: TetR family transcriptional regulators (TFRs) are widely distributed in bacteria and archaea, and the first discovered TFR was confirmed to control the expression of tetracycline efflux pump in Escherichia coli. TFRs can bind DNAs and ligands. Small molecule ligands can induce conformational changes of TFRs, inhibiting or promoting TFRs to control target gene expression. Currently, TFRs have a wide variety of ligands, including carbohydrates, proteins, fatty acids and their derivatives, metal ions, and so on. Due to the diversity of ligands, TFRs regulate a wide range of physiological processes, from basic carbon metabolism and nitrogen metabolism to quorum sensing and antibiotic biosynthesis. On the basis of the recent studies in our laboratory and the literature, we review here the regulatory mechanism mediated by ligands of TFRs in primary and secondary metabolism, as well as the application of ligands for TFRs in the development of gene route and the activation of antibiotic biosynthesis.
Keywords: bacteriaarchaeaTetR family transcription factorsligandfeedback regulationfeed-forward control
原核生物基因组中存在着诸多转录调控因子家族,其中以TetR家族转录调控因子(TetR family transcriptional regulators,TFRs) 数量最多[1]。“TFR”源自四环素抗性阻遏蛋白(Tet repressor, TetR),它是第一个被表征的TetR家族成员[1-2]。TFRs广泛存在于细菌和古菌中,如常见的大肠杆菌Escherichia coli M.、放线菌中的结核分枝杆菌Mycobacterium tuberculosis Z.和抗生素产生菌,以及古菌中的硫化叶菌Sulfolobus B.等,它们参与调控诸多生理代谢活动,如生物膜形成、形态分化、抗生素生物合成、细胞通讯等[2-3]。
TFRs包含N端DNA结合结构域(DNA binding domain,DBD) 与C端配体结合结构域(Ligand binding domain,LBD),具有DNA结合和配体响应的双重能力,这是它们发挥别构调控的结构基础[4]。TFRs LBD的一级序列差异很大,暗示TFRs配体的结构多样性。已知的TFRs配体种类繁多,涵盖糖类、蛋白质、脂肪酸及其衍生物、金属离子等[2]。这种配体多样性使TFRs可以调节的生理过程非常多,包括碳氮代谢、生物膜的形成、群体感应、抗生素的生物合成等[4]。本文主要综述了TFRs配体的研究进展,从配体介导调控角度,揭示配体诱导TFRs调控一系列细胞生理过程的分子机制,并阐述了TFRs配体在基因线路的开发和抗生素合成的激活等方面的应用研究。
1 TFRs配体的研究技术配体作用TFRs LBD导致其DBD构象变化,以“开关”形式激活或阻遏TFRs结合靶点,动态调节靶基因的表达[5]。配体对TFRs发挥别构调控作用至关重要,但其结构多样性与复杂性无疑增加了筛选与鉴别的难度。通常TFRs调控邻近基因表达,其靶基因参与的代谢途径的中间产物可作为TFRs潜在的配体,如AccR的配体[6],但这一方法并不适用于大多数的TFRs。
目前系统基因组学[7]、结构生物学[8]和化合物生物合成途径预测[6]是寻找潜在配体的有效手段。借助于差示扫描荧光法(Differential scanning fluorimetry,DSF) 可实现对TFRs配体的快速、高通量筛选[9]。而凝胶迁移实验(Electrophoretic mobility shift assay,EMSA)[10]、表面等离子共振技术(Surface plasmon resonance,SPR)、等温滴定量热法(Isothermal titration calorimetry,ITC)、荧光偏振法(Fluorescence polarization,FP)、生物膜层干涉技术(Biolayer interferometry,BLI)[11-12]、电喷雾质谱法(Electrospray ionization mass spectrometry,ESI-MS)[13]等分析方法,也为配体与TFRs结合亲和力的表征提供了技术保障。
2 TFRs配体的作用方式蛋白晶体学研究证实小分子配体至少以3种方式进入TFRs LBD的内腔疏水区域(图 1)[2]。(1) “侧进入”:与TFRs二聚化界面侧边入口的氨基酸残基作用进入腔体,例如ActR[14]和FadRSa[15];(2) “前进入”:与TFRs二聚化界面前端的氨基酸残基结合进入内腔,如SimR[16];(3) “顶进入”:与位于TFRs二聚体顶端的氨基酸残基作用进入内腔,如DesT[17]。有趣的是,谷氨酸棒状杆菌中TFR RolR二聚体蛋白可直接包覆其配体间苯二酚,但尚未发现明显的配体进入位点[18]。此外,一些TFRs的配体结合腔中存在多个小分子作用位点,例如SimR[16]和QacR[19],特别是作为多药物输出泵的调控因子QacR,可响应超过16种不同的配体,这进一步突显了其介导调控的复杂性[19]。
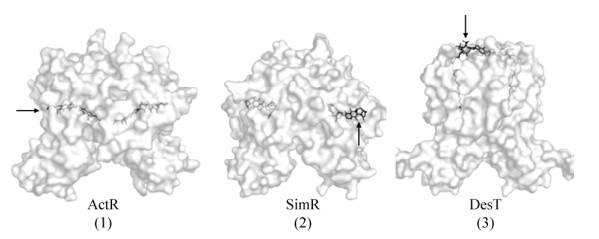 |
| 图 1 TFRs配体不同进入位点示意图[2] Fig. 1 The different entry points of TFRs for ligands[2]. Arrows indicate entry points. (1) "Side entry"; (2) "Front entry"; (3) "Top entry". |
| 图选项 |
3 TFRs配体介导的初级代谢微生物通过各种初级代谢活动合成自身生长所需的物质和能量,而这些生理过程通常受到诸多TFRs及其配体的协同调控,TFRs通过应答小分子配体对不同生理反应实施动态控制,维持微生物的正常生长繁殖(表 1)。
表 1 TFRs配体作用靶点及其对初级代谢的影响Table 1 The targets of ligands for TFRs and their effects on primary metabolism
| Ligands | Targets | Effects | References |
| Citrate-Mg2+ | AcnR | TCA cycle | [20-21] |
| 3-oxocholest-4-en-26-oyl-CoA (3OCh-CoA), 3-oxo-23, 24-bisnorchol-4-en-22-oyl-CoA (4-BNC-CoA) | KstR | Cholesterol degradation | [22] |
| Dihydroxyacetone (Dha) | DhaS-DhaQ | Glycerin degradation | [23] |
| Niacin (NA), 6-hydroxynicotinic acid (6HNA) | NicS | Niacin metabolism | [24] |
| Resorcinol, hydroxyquinol | RolR | Resorcinol degradation | [25] |
| Phenylacetyl-CoA | PaaR | Phenylacetic acid degradation | [26] |
| Palmityl-CoA | Fad35R | Fatty acid biosynthesis | [27] |
| Unsaturated fatty acid (UFA)-CoA, saturated fatty acid (SFA)-CoA | DesT, FabR | Fatty acid metabolism | [28-30] |
| Long-chain acyl-CoAs (C16:0, C16:1, C18:0, C18:1) | FabR_ec, FadR_she | Fatty acid biosynthesis | [31] |
| Lauroyl-CoA, long-chain acyl-CoAs (C10:0, C12:0, C14:0, C18:0) | FadRSa | Fatty acid metabolism | [15] |
| Adenylylated glnK | AmtR | Nitrogen metabolism | [32] |
| Heme | HrtR | Heme efflux | [33] |
| Uracil | RutR | Pyrimidine metabolism | [34] |
| S-adenosylhomocysteine (SAH) | McbR | Sulfur transport and metabolism | [35] |
| Agmatine | AguR | Agmatine utilization | [36] |
| Cyclic di-AMP | DarR | Biofilm formation | [37] |
表选项
3.1 基础碳代谢TFRs应答特定配体实现对基础碳代谢的调控。谷氨酸棒杆菌Corynebacterium glutamicum K. 的acn编码的乌头酸酶可将柠檬酸异构化,是柠檬酸循环(TCA cycle) 的关键节点[20],García-Nafría等[21]报道柠檬酸盐能够抑制TFR AcnR与acn启动子的结合。胆固醇是结核分枝杆菌在感染期的主要碳源,TFR KstR抑制相关酶编码基因的表达,控制胆固醇的降解,而降解产物3OCh-CoA和4-BNC-CoA能够介导KstR的调控[22]。少数TFRs应答小分子配体需要辅助蛋白的参与,例如乳酸乳球菌Lactococcus lactis L. TFR DhaS调控甘油降解途径中二羟基丙酮(Dihydroxyacetone,Dha) 激酶的表达,该激酶的同源物DhaQ与DhaS形成的复合物在Dha存在时会激活dha操纵子的转录[23]。
TFRs参与的碳代谢分解途径中,TFR-DNA复合物可以感知胞内底物或中间体水平,形成闭环的控制回路。例如恶臭假单胞菌Pseudomonas putida T.的TFR NicS作用pa启动子区抑制烟酸(Nicotinic acid, NA) 转化为6-羟基烟酸(6-hydroxynicotinic acid, 6HNA),而NA和6HNA存在时能够诱导NicS与靶点解离,促进NA的分解[24]。TFR RolR抑制基因簇rol调控谷氨酸棒杆菌的间苯二酚分解,胞内积累的间苯二酚和代谢中间体羟基喹诺酚竞争性结合RolR,激活rol基因转录,加速间苯二酚代谢[25]。始旋链霉菌Streptomyces pristinaespiralis R.的TFR PaaR调控苯乙酸的降解途径,苯乙酰-CoA是介导其调控的配体[26]。上述研究表明,当碳代谢途径的底物、中间体或产物积累到浓度阈值时,可结合TFR,促进或抑制其靶基因转录,形成局部的自动控制回路,以浓度依赖方式实现对代谢物的动态调节。
3.2 脂肪酸生物合成与降解FadR是一种脂肪酰基-CoA依赖性转录调控因子,控制脂肪酸生物合成与降解途径中的多种基因。Anand等[27]报道脂肪酸代谢显著影响结核分枝杆菌的致病性,作为FadR的同系物,TFR Fad35R调控酰基-CoA合成酶(Fad) 的表达,棕榈酰-CoA被证实介导这一调控过程,从而控制脂肪酸活化。大肠杆菌的FabR通过调控fabA和fabB的表达水平影响不饱和脂肪酸的生物合成[28],而铜绿假单胞菌Pseudomonas aeruginosa S.的DesT结合desC和desB启动子实现酰基链的去饱和[29],二者调控脂肪酸代谢的不同途径,它们与各自靶点的结合均能被不饱和酰基-CoA增强而被饱和酰基-CoA抑制[30],说明其调控功能存在保守性。沙门氏菌Shewanella oneidensis V.的FadR_she结合fabA调控脂肪酸的生物合成,而大肠杆菌FabR_ec作为其同源蛋白也可以跨种调控fadA,二者对fadA的结合均能被C16和C18脂肪酰基-CoA抑制[31],表明脂肪酰基-CoA对FadR及其同系物的介导调控可能普遍存在于细菌的脂肪酸代谢中。
古菌具有典型的细菌样转录调节因子[38]。FadRSa是TetR家族的古菌成员,它与芽孢杆菌Bacillus C. FadR表现出较高的序列和功能相似性(图 2)。例如在嗜酸热硫化叶菌Sulfolobus acidocaldarius B.中,TFR FadRSa抑制Saci_1103-Saci_1126基因簇控制脂肪酸代谢,月桂酰-CoA和多种长链脂肪酰基-CoA是其配体[15]。FadRSa同系物可能广泛存在于古菌中(图 3),暗示着它们功能上的保守性。
 |
| 图 2 嗜酸热硫化叶菌FadRSa与芽孢杆菌中其同源蛋白的氨基酸序列比对 Fig. 2 Amino acid sequence alignment of FadRSa in S. acidocaldarius and its homologous proteins in Bacillus spp.. S. acidocaldarius FadRSa (GenBank accession number: AAY80459); Bacillus subtilis ETK61_15655 (GenBank accession number: QAW34163); Bacillus halotolerans FLQ13_01120 (GenBank accession number: QDK66354); Bacillus licheniformis BHT94_05605 (GenBank accession number: OLQ51560); Bacillus vallismortis BV11031_03720 (GenBank accession number: QAV07766); Bacillus tequilensis G4P54_14545 (GenBank accession number: QIW80929); Bacillus mojavensis HC660_27320 (GenBank accession number: QJC97205); Bacillus atrophaeus D068_cds28420 (GenBank accession number: AKL85870); Bacillus nakamurai AXI59_12940 (GenBank accession number: KXZ21325); Bacillus amyloliquefaciens LL3_02942 (GenBank accession number: AEB64473). |
| 图选项 |
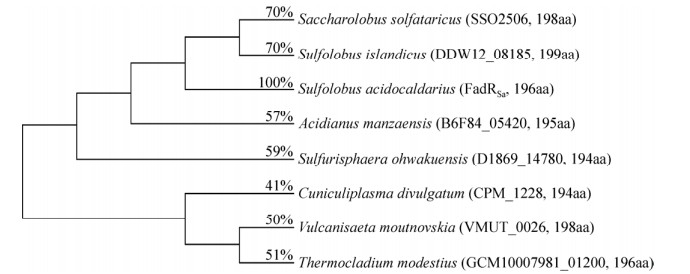 |
| 图 3 古菌中FadRSa及其同系物的进化树分析 Fig. 3 Phylogenetic tree analysis of FadRSa and its homologs in archaea. Percentages represent the identities between FadRSa and its homologs. Sulfolobus acidocaldarius FadRSa (GenBank accession number: AAY80459); Saccharolobus solfataricus SSO2506 (GenBank accession number: AAK42639); Sulfolobus islandicus DDW12_08185 (GenBank accession number: PVU77113); Acidianus manzaensis B6F84_05420 (GenBank accession number: ARM75525); Sulfurisphaera ohwakuensis D1869_14780 (GenBank accession number: QGR18311); Cuniculiplasma divulgatum CPM_1228 (GenBank accession number: SJK85035); Vulcanisaeta moutnovskia VMUT_0026 (GenBank accession number: ADY00243); Thermocladium modestius GCM10007981_01200 (GenBank accession number: GGP19057). |
| 图选项 |
3.3 其他基础代谢除了参与上述碳基础代谢和脂肪酸代谢,TFRs配体还控制氮代谢、嘧啶代谢、氨基酸代谢、生物膜形成等诸多生理活动。TFR AmtR是棒状杆菌氮代谢的主要调控因子,腺苷酸化的GlnK蛋白可以诱导其与靶点的解离[32]。Lechardeur等[33]发现HrtR可以响应血红素,调控乳酸乳球菌血红素的外排。RutR是大肠杆菌嘧啶代谢的主要调节剂,配体是尿嘧啶[34],其同源蛋白TFR PydR同样控制恶臭假单胞菌嘧啶的降解,并且RutR应答尿嘧啶的氨基酸残基在PydR中保守,表明PydR可能同样对尿嘧啶敏感[39]。Rey等[35]证实McbR是谷氨酸棒杆菌中涉及硫代谢的全局调节因子,直接抑制至少45个基因的转录,这些基因涉及(S-腺苷) 蛋氨酸和半胱氨酸合成、硫酸盐还原、含硫化合物吸收和利用等,S-腺苷同型半胱氨酸阻止McbR结合靶点,调控大分子硫的运输和代谢。
胍丁胺是铜绿假单胞菌精氨酸降解的中间体,研究表明TFR AguR结合aguAB启动子调控胍丁胺的利用,反过来又可被胍丁胺诱导,序列分析显示AguR在假单胞菌属许多成员中都是保守的[36]。细胞中存在一类可扩散的信号分子,在纳摩尔水平即可诱导TFRs实现对多种生理活动的灵敏调控,例如耻垢分枝杆菌Mycobacterium smegmatis T.中第二信使环状di-AMP (Cyclic di-AMP) 介导TFR DarR,实现对细菌的生物膜形成和运动的控制[37]。
4 TFRs配体介导的次级代谢微生物凭借自身复杂的次级代谢系统,可以合成诸多生物活性显著、结构多样的抗生素。在抗生素生物合成时,通常其前体、中间体和终产物以及信号分子充当配体介导TFR的调控(表 2)。
表 2 TFRs配体作用靶点及其对次级代谢的影响Table 2 The targets of ligands for TFRs and their effects on secondary metabolism
| Ligands | Targets | Effects | References |
| Methylmalonic acid | PccD | Erythromycin biosynthesis | [40] |
| Methylcrotonyl, propionyl, acetyl-CoA | AccR | Avermectin biosynthesis | [6] |
| 2, 3-dehydro-UWM6 (DHU), dehydrorabelomycin (DHR), jadomycin B (JdB) | JadR* | JdB biosynthesis | [41] |
| C-5–O-B1 | AveT | Avermectin biosynthesis | [42] |
| 4-dihydro-9-hydroxy-1-methyl-10-oxo -3-H-naphtho-[2, 3-c]-pyran-3-(S)-acetic acid ((S)-DNPA), actinorhodin (Act) | ActR | Act efflux | [14, 43] |
| Cezomycin, calcimycin | CalR3 | Calcimycin biosynthesis | [44] |
| Heptaene | AtrA | Lidamycin biosynthesis | [45] |
| Chlorothricin, demethylsalicycloyl chlorothricin, deschloro-chlorothricin | CHlF1 | Chlorothricin biosynthesis | [46] |
| Act, undecylprodigiosin (Red), JdB | ScbR2 | Cryptic polyketide (CPK) and Red biosynthesis | [47-48] |
| Chloramphenicol (Cm), JdB | JadR2 | JdB and Cm biosynthesis | [5] |
| Tetracycline | TetR | Tetracycline efflux | [2] |
| Simocyclinone C4, simocyclinone D8 (SD8) | SimR | SD8 efflux | [49] |
| Rifamycin B | RifQ | Rifamycin B efflux | [50] |
| Virginiamycin S | VarR | Virginiamycin S efflux | [51] |
| A-factor | ArpA | Streptomycin biosynthesis | [52-53] |
| 2-(1?-hydroxy-6-methylheptyl)-3-hydroxymethylbutanolide (SCB1), 2-(1?-hydroxyoctyl)-3-hydroxymethylbutanolide (SCB2), 2-(1?-hydroxy-6?-methyloctyl)-3-hydroxymethylbutanolide (SCB3) | ScbR | Act and Red biosynthesis | [54-55] |
| SVB1 | JadR3 | JdB biosynthesis | [56] |
| Avenolide | AvaR1, AvaR2 | Avermectin biosynthesis | [57-58] |
| ppGpp, GTP | XdhR | Purine salvage pathway | [59] |
| 14-membered macrolides | MphR | Macrolides biosynthesis | [60] |
表选项
4.1 抗生素生物合成前体微生物的初级代谢除了维持自身生长需要,还为其次级代谢物的合成提供了丰富的前体[5]。研究表明抗生素生物合成前体可充当调控配体,影响某些特定的代谢通路。例如在红色糖多孢菌Saccharopolyspora erythraea W.中,作为红霉素生物合成的羧酸类前体,甲基丙二酸介导TFR PccD对丙酰-CoA羧化酶基因的调控,影响甲基丙二酰- CoA的胞内供应[40]。而近期Lyu等[6]发现阿维链霉菌Streptomyces avermitilis B.的TFR AccR抑制酰基-CoA羧化酶基因accD1A1的转录,影响胞内5种酰基-CoA的吸收,而甲基巴豆酰、丙酰和乙酰-CoA介导这一调控过程。尽管这3种酰基- CoA不能直接参与阿维菌素生物合成,却是其他聚酮抗生素生物合成的前体。
4.2 抗生素生物合成中间体代谢中间体常以前馈或反馈形式介导TFR对靶点的控制,避免物质的过度积累,稳定抗生素生物合成。Zhang等[41]报道JadR*结合杰多霉素B (Jadomycin B, JdB) 生物合成簇内基因jadY等抑制其生物合成,早期中间体DHU和DHR抑制JadR*对jadY启动子的控制。阿维链霉菌中,Liu等[42]发现阿维菌素B1生物合成后期中间体C-5–O-B1的过多积累会阻遏TFR AveT结合靶基因。这种相似的诱导调控机制可能普遍存在于产抗生素的放线菌中。
Nodwell团队[14]报道在天蓝色链霉菌Streptomyces coelicolor M.放线紫红素(Actinorhodin,Act) 生物合成途径中,中间体(S)-DNPA和Act均能影响ActR对Act外排的调控;而Xu等[43]深入研究后发现Act输出存在两步调控,在Act浓度相对较低的合成前期,(S)-DNPA主要介导输出基因的表达,而随着Act积累,Act便触发持续的外排泵表达。在其他放线菌中同样不乏代谢中间体介导TFR的报道,如钙霉素的中间体色唑霉素[44]和力达霉素的中间体Heptaene[45]。这种以配体浓度前馈或反馈调节TFR的方式,平衡代谢中间体与抗生素合成,是很多次级代谢产物生物合成调控的普遍规律。
4.3 内源性抗生素尽管高浓度的抗生素被用于抵御环境微生物,但其低浓度的抗生素可作为化学诱导物诱发细胞的多种代谢变化[5]。某些放线菌中,抗生素生物合成基因簇内存在途径特异性调控因子,它们可以响应内源抗生素,动态调控抗生素簇内生物合成基因的表达。
Xu等[47]发现天蓝色链霉菌TFR ScbR2抑制簇内激活剂KasO负调隐性Ⅰ型聚酮(Cryptic polyketide,CPK) 的生产,Act和十一烷基灵菌红素(Undecylprodigiosin, Red) 竞争性结合ScbR2激活kasO,从而开启CPK的生物合成。同样地,委内瑞拉链霉菌Streptomyces venezuelae E. 的TFR JadR2遏制JadR1调控JdB生物合成,JdB的存在会干扰JadR2对JadR1的抑制,刺激JdB生产;JadR2还可以抑制JadR1激活氯霉素(Chloramphenicol, Cm) 生物合成,而Cm的积累会反馈抑制JadR2的DNA结合活性。表明不同抗生素可以介导同一TFR,实现对自身生物合成的动态调控,而这种调控模式广泛存在于抗生素产生菌中[5] (图 4)。
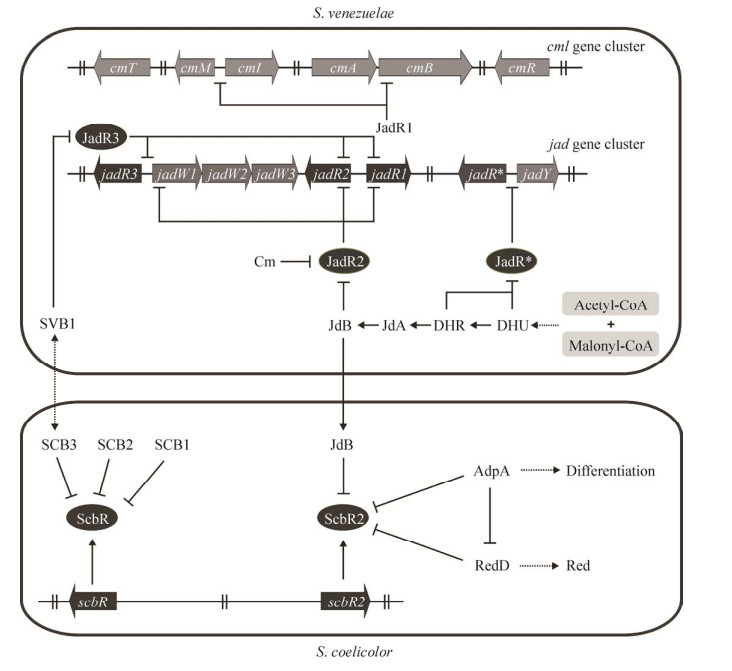 |
| 图 4 委内瑞拉链霉菌和天蓝色链霉菌中配体介导TFRs的调控[5] Fig. 4 Ligand-mediated regulation of TFRs in S. venezuelae and S. coelicolor[5]. JadR2, JadR3, JadR*, the TetR family transcriptional regulator in S. venezuelae; ScbR, ScbR2, the TetR family transcriptional regulator in S. coelicolor. JdA, Jadomycin A; DHU, 2, 3-dehydro-UWM6; DHR, dehydrorabelomycin; SCB1, 2-(1?-hydroxy-6-methylheptyl)-3- hydroxymethylbutanolide; SCB2, 2-(1?-hydroxyoctyl)-3-hydroxymethylbu-tanolide; SCB3, 2-(1?-hydroxy-6?-methyloctyl)- 3-hydroxymethylbutanolide. |
| 图选项 |
微生物具备不同抗性系统来抵御毒性分子(毒物和抗生素等) 侵害。研究表明许多TFRs可以应答抗生素调控细胞的耐药性,如TFR家族的首位成员TetR,可以响应四环素并调控其外排[2]。Le等[49]发现TFR SimR作用外排泵基因simX实现抗生链霉菌Streptomyces antibioticus W. 对Simocyclinone D8 (SD8) 的耐药性,而SD8及其中间体simocyclinone C4能够阻遏SimR对simX的控制;而随后的研究则从结构水平揭示了SD8介导SimR调控的更多原子细节[16]。近期,笔者实验室发现林可链霉菌Streptomyces lincolnensis M.的TFR SLCG_2919调控ATP/GTP结合蛋白基因SLCG_2920,过表达该基因可提高菌株对林可霉素抗性,推测林可霉素可能介导SLCG_2919的调控[61]。类似功能的抗生素还有很多,如利福霉素B[50]和弗吉尼亚霉素S[51]等。同TetR/tetA外排系统一样,这些抗性基因受TFRs调控,当胞内抗生素积累到毒性阈值,便激活被TFRs抑制的泵基因的表达,实现抗生素外排与其生物合成的偶联,这或许是许多抗生素生产中的共同特征。
4.4 外源性抗生素外源性抗生素具有比内源性抗生素更为复杂的介导机制,它能够实现跨种属的诱导调控,是细菌进化的一种重要生存策略。例如委内瑞拉链霉菌产生的JdB可诱导天蓝色链霉菌产生复杂的生理反应[48] (图 4)。AdpA是形态分化和次级代谢的主要调节剂,TFR ScbR2通过结合redD和adpA启动子,直接调控Red生产和菌株形态分化,同时AdpA还直接调控redD的表达,ScbR2响应外源JdB,在ScbR2-AdpA-RedD间形成前馈环,控制天蓝色链霉菌的形态分化和Red生物合成[48, 62]。
4.5 信号分子灰色链霉菌Streptomyces griseus K. A因子(A-factor) 是细菌中第一个被表征的γ-丁内酯(Gamma butyrolactone, GBL) 类信号分子[52],A因子诱导一系列级联反应影响链霉素产生和菌株形态分化,深入研究发现这一级联反应需要A因子受体ArpA、多效调节因子AdpA和簇内激活因子StrR参与[53]。得益于A因子研究的启发,更多的激素样信号分子被发现,例如天蓝色链霉菌中鉴定的SCB1、SCB2和SCB3[54-55]、委内瑞拉链霉菌中TFR JadR3的配体SVB1[56]等。有趣的是,SVB1和SCB3结构相同,继续研究证实SVB1/SCB3可作为种间信号,ScbR和JadR3响应彼此释放的外源性SVB1/SCB3,调控内源抗生素的生物合成[56](图 4)。越来越多的相关研究不断加深人们对信号分子介导抗生素生物合成级联调控机制的认识。
与GBL功能类似的呋喃霉素(Methylenomycin furan,MMF)、ppGpp、GTP等,同样可作为TFR的诱导剂。MMFs特异地诱导天蓝色链霉菌次甲霉素的生物合成[63]。天蓝色链霉菌中ppGpp和GTP介导TFR XdhR对嘌呤挽救途径的调控[59]。这种纳摩尔浓度便可发挥作用和可扩散的信号分子,无疑增加了细菌调控的灵敏性和机动性,同时也增强了其对多变生境的适应能力。
5 TFRs配体的开发与应用5.1 基因线路的开发TFRs靶基因和配体可被用作基本的生物零件,设计构建出像Tet-Off/On系统的基因线路,再利用配体去激活或关闭下游基因表达。例如利用TFR PhlF开发的DAPG-Off/On触发开关系统可调控酿酒酵母Saccharomyces cerevisiae H.中报告基因表达。2, 4-二乙酰基间苯三酚(2, 4-diacetylphloroglucinol, DAPG) 可以诱导PhlF与靶点(phlO) 的解离。DAPG-Off系统中,与多聚病毒激活蛋白结构域融合的PhlF激活报告基因表达,当添加DAPG时,该基因的表达被关闭;与DAPG-Off效果相反的DAPG-On系统,DAPG存在会开启报告基因表达[64]。同样基于TFR CamR配体樟脑(Camphor) 开发的Camphor-Off系统应用于激活无樟脑体系中报告基因的表达[65]。
除了触发开关系统,基于TFRs及其配体开发的自调控基因线路,可实时监测和动态调控目标产物的生物合成,为提高“细胞工厂”的生产力及突破微生物天然产物生物合成的关键瓶颈提供了可能。例如基于TFR MphR定向进化构建的基因线路,显著提高了MphR对大环内酯类抗生素的敏感性,可应用于大环内酯类抗生素和其结构类似物产生菌的快速筛选[60];基于丙二酰-CoA开发的基因线路能够自反馈调节相关代谢物流量、平衡胞内丙二酰-CoA代谢池[66];利用大肠杆菌FadR响应酰基-CoA这一特性,在酿酒酵母中构建与基因过表达文库相结合的基因线路,不仅可以鉴定出增强酰基-CoA水平的基因靶点,还大幅提高了脂肪酸衍生物脂肪醇的产量[67]。基于合成生物学理念,利用TFR及其配体等基本的生物零件开发出可应用的基因线路,俨然成为突破细菌天然产物生物合成瓶颈、提高其代谢效价的一种重要手段。
5.2 抗生素生物合成的激活GBLs类信号分子可以作为抗生素生产的诱导剂[5],但受限于天然GBLs的产量,人工合成的GBLs及其结构类似物是天然GBLs有效替代品。研究发现添加合成的SCB1可促进天蓝色链霉菌Act的产生[68]。同样地,在吸水链球菌Streptomyces hygroscopicus J.和螺旋霉素链霉菌Streptomyces spiramyceticus Y.中分别添加化学合成GBL类似物1, 4-丁内酯,促进了井冈霉素和双螺旋霉素的生物合成[69-70]。因此,外源添加信号分子及其结构类似物是一种改善抗生素产量的有效策略[53]。
6 总结与展望本文主要阐述了近年来TFRs配体的研究进展,旨在更好地了解配体介导TFRs调控的分子机制,为应用TFRs配体提高活性天然产物的代谢效价提供一定的参考。笔者实验室一直致力于红色糖多孢菌中TFRs的调控功能解析[71-74],构建了基于TFRs的微型调控网络,筛选鉴定出多个TFRs的小分子配体,并初步揭示了配体诱导TFRs调控的分子机制。由于TFRs复杂的作用机制和其配体种类的多样性,仍有大量TFRs配体没有被发现。例如作为多种酶促反应必要的辅助因子,生物素的生物合成受到TFR BioQ的调控,但目前还没有发现BioQ的调控配体[75]。大肠杆菌的ComR和肺炎链球菌Streptococcus pneumoniae K.的SczA都能感应金属离子,但这些相互作用的分子细节尚不清楚[76-77]。TFR ScbR2不仅感应内源配体Act和Red,还能被外源的JdB诱导[48];天蓝色链霉菌的ScbR和委内瑞拉链霉菌的JadR3响应彼此释放的外源性SVB1/SCB3,调控自身抗生素的生物合成[56]。这些外源分子或其结构类似物同样能诱导TFRs调控,这无疑增加了TFRs配体识别的难度。放线菌基因组存在众多的隐性合成基因簇,这些尚未表征的基因簇是新型活性天然产物的重要来源[78],配体分子可以充当诱导物,激活隐性基因簇,进一步丰富活性天然产物的多样性[53, 63]。
寻找配体是TFRs功能研究中非常重要的一环,建立完善的TFR配体库将是配体筛选的关键。近年来,研究人员利用系统基因组学[7]、结构生物学[8]、化合物生物合成途径预测[6]等手段鉴定了很多TFRs配体,但配体如何诱导TFRs产生别构效应、怎样精确介导其调控仍研究较少,加之配体介导调控的网络尚不清楚;不同配体介导同一TFR或同一配体诱导不同TFRs的调控中,是否存在竞争性互作的优先级、其信号如何实现级联转导等也需要深入研究。阐明这些配体在次级代谢中的作用,对于改造生物合成途径提高抗生素产量具有重要的现实意义。
利用配体诱导TFRs产生的别构调节效应,不但可以开发出具备不同功能模块的基因线路,实现对报告基因表达的人为控制(如Tet-Off/On触发开关) 和生物传感器系统的自动调整(如反馈调节基因线路),而且能促进医用抗生素和生物燃料等次级产物的生产,同时也有助于激活隐性抗生素生物合成途径。如何突破配体应用的瓶颈,探索配体开发的新途径,将配体研究成果应用到合成生物学基因线路的开发、隐性基因簇的激活以及微生物药物的高效制造等方面将是未来的研究热点。
参考文献
| [1] | Ramos JL, Martínez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev, 2005, 69(2): 326-356. |
| [2] | Cuthbertson L, Nodwell JR. The TetR family of regulators. Microbiol Mol Biol Rev, 2013, 77(3): 440-475. |
| [3] | Balhana RJ, Singla A, Sikder MH, et al. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics, 2015, 16(1): 479-490. DOI:10.1186/s12864-015-1696-9 |
| [4] | Yu Z, Reichheld SE, Savchenko A, et al. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J Mol Biol, 2010, 400(4): 847-864. DOI:10.1016/j.jmb.2010.05.062 |
| [5] | Niu GQ, Chater KF, Tian YQ, et al. Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol Rev, 2016, 40(4): 554-573. DOI:10.1093/femsre/fuw012 |
| [6] | Lyu MY, Cheng YQ, Han X, et al. AccR, a TetR family transcriptional repressor, coordinates short-chain acyl coenzyme A homeostasis in Streptomyces avermitilis. Appl Environ Microbiol, 2020, 86(12): e00508-20. |
| [7] | Zhang H, White-Phillip JA, Melan?on 3rd CE, et al. Elucidation of the kijanimicin gene cluster: insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J Am Chem Soc, 2007, 129(47): 14670-14683. DOI:10.1021/ja0744854 |
| [8] | Frénois F, Engohang-Ndong J, Locht C, et al. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol Cell, 2004, 16(2): 301-307. DOI:10.1016/j.molcel.2004.09.020 |
| [9] | McClure SM, Ahl PL, Blue JT. High throughput differential scanning fluorimetry (DSF) formulation screening with complementary dyes to assess protein unfolding and aggregation in presence of surfactants. Pharm Res, 2018, 35(4): 81-90. DOI:10.1007/s11095-018-2361-1 |
| [10] | Aung KM, New SY, Hong SZ, et al. Studying forkhead box protein A1-DNA interaction and ligand inhibition using gold nanoparticles, electrophoretic mobility shift assay, and fluorescence anisotropy. Anal Biochem, 2014, 448: 95-104. DOI:10.1016/j.ab.2013.11.017 |
| [11] | Du X, Li Y, Xia YL, et al. Insights into protein-ligand interactions: mechanisms, models, and methods. Int J Mol Sci, 2016, 17(2): 144-177. DOI:10.3390/ijms17020144 |
| [12] | Kairys V, Baranauskiene L, Kazlauskiene M, et al. Binding affinity in drug design: experimental and computational techniques. Expert Opin Drug Discov, 2019, 14(8): 755-768. DOI:10.1080/17460441.2019.1623202 |
| [13] | Pacholarz KJ, Garlish RA, Taylor RJ, et al. Mass spectrometry based tools to investigate protein-ligand interactions for drug discovery. Chem Soc Rev, 2012, 41(11): 4335-4355. DOI:10.1039/c2cs35035a |
| [14] | Willems AR, Tahlan K, Taguchi T, et al. Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J Mol Biol, 2008, 376(5): 1377-1387. DOI:10.1016/j.jmb.2007.12.061 |
| [15] | Wang K, Sybers D, Maklad HR, et al. A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nat Commun, 2019, 10(1): 1542-1557. DOI:10.1038/s41467-019-09479-1 |
| [16] | Le TBK, Stevenson CEM, Fiedler H, et al. Structures of the TetR-like simocyclinone efflux pump repressor, SimR, and the mechanism of ligand-mediated derepression. J Mol Biol, 2011, 408(1): 40-56. DOI:10.1016/j.jmb.2011.02.035 |
| [17] | Miller DJ, Zhang YM, Subramanian C, et al. Structural basis for the transcriptional regulation of membrane lipid homeostasis. Nat Struct Mol Biol, 2010, 17(8): 971-975. DOI:10.1038/nsmb.1847 |
| [18] | Li DF, Zhang N, Hou YJ, et al. Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator. PLoS One, 2011, 6(5): e19529-38. DOI:10.1371/journal.pone.0019529 |
| [19] | Takeuchi K, Imai M, Shimada I. Conformational equilibrium defines the variable induction of the multidrug-binding transcriptional repressor QacR. Proc Natl Acad Sci USA, 2019, 116(40): 19963-19972. DOI:10.1073/pnas.1906129116 |
| [20] | Krug A, Wendisch VF, Bott M. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J Biol Chem, 2005, 280(1): 585-595. DOI:10.1074/jbc.M408271200 |
| [21] | García-Nafría J, Baumgart M, Turkenburg JP, et al. Crystal and solution studies reveal that the transcriptional regulator AcnR of Corynebacterium glutamicum is regulated by citrate-Mg2+ binding to a non-canonical pocket. J Biol Chem, 2013, 288(22): 15800-15812. DOI:10.1074/jbc.M113.462440 |
| [22] | Ho NA, Dawes SS, Crowe AM, et al. The structure of the transcriptional repressor KstR in complex with CoA thioester cholesterol metabolites sheds light on the regulation of cholesterol catabolism in Mycobacterium tuberculosis. J Biol Chem, 2016, 291(14): 7256-7266. DOI:10.1074/jbc.M115.707760 |
| [23] | Christen S, Srinivas A, B?hler P, et al. Regulation of the Dha operon of Lactococcus lactis: a deviation from the rule followed by the TetR family of transcription regulators. J Biol Chem, 2006, 281(32): 23129-23137. DOI:10.1074/jbc.M603486200 |
| [24] | Jimenez JI, Juarez JF, Garcia JL, et al. A finely tuned regulatory circuit of the nicotinic acid degradation pathway in Pseudomonas putida. Environ Microbiol, 2011, 13(7): 1718-1732. DOI:10.1111/j.1462-2920.2011.02471.x |
| [25] | Li T, Zhao KX, Huang Y, et al. The TetR-type transcriptional repressor RolR from Corynebacterium glutamicum regulates resorcinol catabolism by binding to a unique operator, rolO. Appl Environ Microbiol, 2012, 78(17): 6009-6016. DOI:10.1128/AEM.01304-12 |
| [26] | Zhao YW, Feng RR, Zheng GS, et al. Involvement of the TetR-type regulator PaaR in the regulation of pristinamycin Ⅰ biosynthesis through an effect on precursor supply in Streptomyces pristinaespiralis. J Bacteriol, 2015, 197(12): 2062-2071. DOI:10.1128/JB.00045-15 |
| [27] | Anand S, Singh VP, Singh AK, et al. Equilibrium binding and kinetic characterization of putative tetracycline repressor family transcription regulator Fad35R from Mycobacterium tuberculosis. FEBS J, 2012, 279(17): 3214-3228. DOI:10.1111/j.1742-4658.2012.08707.x |
| [28] | Yang J, Fang Y, Wang JL, et al. Deletion of regulator-encoding genes fadR, fabR and iclR to increase L-threonine production in Escherichia coli. Appl Microbiol Biotechnol, 2019, 103(11): 4549-4564. DOI:10.1007/s00253-019-09818-8 |
| [29] | Subramanian C, Rock CO, Zhang YM. DesT coordinates the expression of anaerobic and aerobic pathways for unsaturated fatty acid biosynthesis in Pseudomonas aeruginosa. J Bacteriol, 2010, 192(1): 280-285. DOI:10.1128/JB.00404-09 |
| [30] | Feng YJ, Cronan JE. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol, 2011, 80(1): 195-218. DOI:10.1111/j.1365-2958.2011.07564.x |
| [31] | Zhang HM, Zheng BW, Gao RS, et al. Binding of Shewanella FadR to the fabA fatty acid biosynthetic gene: implications for contraction of the fad regulon. Protein Cell, 2015, 6(9): 667-679. DOI:10.1007/s13238-015-0172-2 |
| [32] | Beckers G, Strosser J, Hildebrandt U, et al. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol Microbiol, 2005, 58(2): 580-595. DOI:10.1111/j.1365-2958.2005.04855.x |
| [33] | Lechardeur D, Cesselin B, Liebl U, et al. Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB-HrtA transporter in Lactococcus lactis. J Biol Chem, 2012, 287(7): 4752-4758. DOI:10.1074/jbc.M111.297531 |
| [34] | Nguyen P, Bervoets I, Maes D, et al. The protein- DNA contacts in RutR?carAB operator complexes. Nucleic Acids Res, 2010, 38(18): 6286-6300. DOI:10.1093/nar/gkq385 |
| [35] | Rey DA, Nentwich SS, Koch DJ, et al. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol Microbiol, 2005, 56(4): 871-887. DOI:10.1111/j.1365-2958.2005.04586.x |
| [36] | Gilbertsen A, Williams B. Development of a Pseudomonas aeruginosa agmatine biosensor. Biosensors (Basel), 2014, 4(4): 387-402. |
| [37] | Zhang L, Li WH, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem, 2013, 288(5): 3085-3096. DOI:10.1074/jbc.M112.428110 |
| [38] | Lemmens L, Maklad HR, Bervoets I, et al. Transcription regulators in archaea: homologies and differences with bacterial regulators. J Mol Biol, 2019, 431(20): 4132-4146. DOI:10.1016/j.jmb.2019.05.045 |
| [39] | Hidese R, Mihara H, Kurihara T, et al. Pseudomonas putida PydR, a RutR-like transcriptional regulator, represses the dihydropyrimidine dehydrogenase gene in the pyrimidine reductive catabolic pathway. J Biochem, 2012, 152(4): 341-346. DOI:10.1093/jb/mvs079 |
| [40] | Xu Z, Wang MM, Ye BC. TetR family transcriptional regulator PccD negatively controls propionyl coenzyme A assimilation in Saccharopolyspora erythraea. J Bacteriol, 2017, 199(20): e00281-17. |
| [41] | Zhang YY, Pan GH, Zou ZZ, et al. JadR*-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol Microbiol, 2013, 90(4): 884-897. DOI:10.1111/mmi.12406 |
| [42] | Liu WS, Zhang QL, Guo J, et al. Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl Environ Microbiol, 2015, 81(15): 5157-5173. DOI:10.1128/AEM.00868-15 |
| [43] | Xu Y, Willems A, Au-Yeung C, et al. A two-step mechanism for the activation of actinorhodin export and resistance in Streptomyces coelicolor. mBio, 2012, 3(5): e00191-12. |
| [44] | Gou LX, Han TS, Wang XX, et al. A novel TetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front Microbiol, 2017, 8: 2371-2380. DOI:10.3389/fmicb.2017.02371 |
| [45] | Li XX, Yu TF, He Q, et al. Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol Microbiol, 2015, 96(6): 1257-1271. DOI:10.1111/mmi.13004 |
| [46] | Li Y, Li JJ, Tian ZH, et al. Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J Biol Chem, 2016, 291(10): 5406-5417. DOI:10.1074/jbc.M115.695874 |
| [47] | Xu GM, Wang J, Wang LQ, et al. "Pseudo" γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem, 2010, 285(35): 27440-27448. DOI:10.1074/jbc.M110.143081 |
| [48] | Wang WS, Ji JJ, Li X, et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc Natl Acad Sci USA, 2014, 111(15): 5688-5693. DOI:10.1073/pnas.1324253111 |
| [49] | Le TBK, Fiedler H, Den Hengst CD, et al. Coupling of the biosynthesis and export of the DNA gyrase inhibitor simocyclinone in Streptomyces antibioticus. Mol Microbiol, 2009, 72(6): 1462-1474. DOI:10.1111/j.1365-2958.2009.06735.x |
| [50] | Lei C, Wang JZ, Liu YY, et al. A feedback regulatory model for RifQ-mediated repression of rifamycin export in Amycolatopsis mediterranei. Microb Cell Fact, 2018, 17(1): 14-22. DOI:10.1186/s12934-018-0863-5 |
| [51] | Namwat W, Lee CK, Kinoshita H, et al. Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae. J Bacteriol, 2001, 183(6): 2025-2031. |
| [52] | Szilágyi M, Márton é, Lukács D, et al. Mutation in afsR leads to A-factor deficiency in Streptomyces griseus B2682. J Mol Microbiol Biotechnol, 2018, 28(5): 216-224. |
| [53] | Kong DK, Wang X, Nie J, et al. Regulation of antibiotic production by signaling molecules in Streptomyces. Front Microbiol, 2019, 10: 2927-2937. |
| [54] | Takano E, Nihira T, Hara Y, et al. Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J Biol Chem, 2000, 275(15): 11010-11016. |
| [55] | Hsiao NH, Nakayama S, Merlo ME, et al. Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem Biol, 2009, 16(9): 951-960. |
| [56] | Zou ZZ, Du DY, Zhang YY, et al. A γ-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol Microbiol, 2014, 94(3): 490-505. |
| [57] | Zhu JY, Chen Z, Li JL, et al. AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front Microbiol, 2017, 8: 2577-2591. |
| [58] | Zhu JY, Su D, Liu WS, et al. AvaR2, a pseudo γ-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol Microbiol, 2016, 102(4): 562-578. |
| [59] | Sivapragasam S, Grove A. Streptomyces coelicolor XdhR is a direct target of (p)ppGpp that controls expression of genes encoding xanthine dehydrogenase to promote purine salvage. Mol Microbiol, 2016, 100(4): 701-718. |
| [60] | Kasey CM, Zerrad M, Li YW, et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology. ACS Synth Biol, 2018, 7(1): 227-239. |
| [61] | Xu YR, Ke ML, Li J, et al. TetR-type regulator SLCG_2919 is a negative regulator of lincomycin biosynthesis in Streptomyces lincolnensis. Appl Environ Microbiol, 2019, 85(1): e02091-18. |
| [62] | Wolański M, Jakimowicz D, Zakrzewska-Czerwińska J. AdpA, key regulator for morphological differentiation regulates bacterial chromosome replication. Open Biol, 2012, 2(7): 120097-120105. |
| [63] | O'Rourke S, Wietzorrek A, Fowler K, et al. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Molecular Microbiology, 2009, 71(3): 763-778. |
| [64] | Ikushima S, Boeke JD. New orthogonal transcriptional switches derived from Tet repressor homologues for Saccharomyces cerevisiae regulated by 2, 4-diacetylphloroglucinol and other ligands. ACS Synth Biol, 2017, 6(3): 497-506. |
| [65] | Ikushima S, Zhao Y, Boeke JD. Development of a tightly controlled off switch for Saccharomyces cerevisiae regulated by camphor, a low-cost natural product. G3 (Bethesda), 2015, 5(10): 1983-1990. |
| [66] | Johnson AO, Gonzalez-Villanueva M, Wong L, et al. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab Eng, 2017, 44: 253-264. |
| [67] | Dabirian Y, Gon?alves Teixeira P, Nielsen J, et al. FadR-based biosensor-assisted screening for genes enhancing fatty acyl-CoA pools in Saccharomyces cerevisiae. ACS Synth Biol, 2019, 8(8): 1788-1800. |
| [68] | Yang YH, Kim TW, Park SH, et al. Cell-free Escherichia coli-based system to screen for quorum-sensing molecules interacting with quorum receptor proteins of Streptomyces coelicolor. Appl Environ Microbiol, 2009, 75(19): 6367-6372. |
| [69] | Tan GY, Bai LQ, Zhong JJ. Exogenous 1, 4-butyrolactone stimulates A-factor-like cascade and validamycin biosynthesis in Streptomyces hygroscopicus 5008. Biotechnol Bioeng, 2013, 110(11): 2984-2993. |
| [70] | Gao XW, Wang YH, Chu J. A preliminary study on the impact of exogenous A-factor analogue 1, 4-butyrolactone on stimulating bitespiramycin biosynthesis. Bioprocess Biosyst Eng, 2019, 42(12): 1903-1913. DOI:10.1007/s00449-019-02184-9 |
| [71] | Wu PP, Pan H, Zhang CM, et al. SACE_3986, a TetR family transcriptional regulator, negatively controls erythromycin biosynthesis in Saccharopolyspora erythraea. J Ind Microbiol Biotechnol, 2014, 41(7): 1159-1167. |
| [72] | Wu H, Chen M, Mao YR, et al. Dissecting and engineering of the TetR family regulator SACE_7301 for enhanced erythromycin production in Saccharopolyspora erythraea. Microb Cell Fact, 2014, 13: 158-166. |
| [73] | Wu H, Wang YS, Yuan L, et al. Inactivation of SACE_3446, a TetR family transcriptional regulator, stimulates erythromycin production in Saccharopolyspora erythraea. Synth Syst Biotechnol, 2016, 1(1): 39-46. |
| [74] | Wu H, Chu ZL, Zhang WX, et al. Transcriptome-guided target identification of the TetR-like regulator SACE_5754 and engineered overproduction of erythromycin in Saccharopolyspora erythraea. J Biol Eng, 2019, 13: 11-22. DOI:10.1186/s13036-018-0135-2 |
| [75] | Brune I, Gotker S, Schneider J, et al. Negative transcriptional control of biotin metabolism genes by the TetR-type regulator BioQ in biotin-auxotrophic Corynebacterium glutamicum ATCC 13032. J Biotechnol, 2012, 159(3): 225-234. |
| [76] | Mermod M, Magnani D, Solioz M, et al. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals, 2012, 25(1): 33-43. |
| [77] | Martin JE, Edmonds KA, Bruce KE, et al. The zinc efflux activator SczA protects Streptococcus pneumoniae serotype 2 D39 from intracellular zinc toxicity. Mol Microbiol, 2017, 104(4): 636-651. |
| [78] | Mao DN, Okada BK, Wu YH, et al. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol, 2018, 45: 156-163. |
