

中国科学院微生物研究所 中国科学院微生物生理与代谢工程重点实验室,北京 100101
收稿日期:2020-10-26;接收日期:2021-02-01;网络出版时间:2021-02-09
基金项目:国家重点研发计划(No. 2018YFA0901400),国家自然科学基金(No. 31970025) 资助
作者简介:于波 ??中国科学院微生物研究所研究员,中国科学院微生物生理与代谢工程重点实验室副主任。2006年于山东大学获得微生物学博士学位,2008–2010年获洪堡基金会资助赴德国从事博士后研究,2010年10月回中国科学院微生物研究所工作。主要研究方向为工业微生物代谢工程与合成生物学技术开发。已发表科研论文60余篇,授权中国发明专利13项。目前兼任北京微生物学会副理事长及工业微生物专业委员会主任、CAS-TWAS生物技术卓越中心执行主任、Applied and Environmental Microbiology期刊编委(Editor abroad).
摘要:代谢调控是构建微生物细胞工厂的重要技术手段。随着合成生物学技术的不断突破,挖掘和人工设计的高质量调控元件大幅度提升了对细胞代谢网络的改造能力;代谢调控研究也已从单基因的静态调控发展到系统水平上的智能精确动态调控。文中简要综述了近30年来代谢途径表达调控技术在代谢工程领域的研究进展。
关键词:代谢工程表达调控合成生物学细胞工厂
Metabolic regulation in constructing microbial cell factories
Yang Liu, Qingxuan Mu, Ya'nan Shi, Bo Yu


CAS Key Laboratory of Microbial Physiological & Metabolic Engineering, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
Received: October 26, 2020; Accepted: February 1, 2021; Published: February 9, 2021
Supported by: National Key Research and Development Program of China (No. 2018YFA0901400), National Natural Science Foundation of China (No. 31970025)
Corresponding author: Bo Yu. Tel/Fax: +86-10-64806132; E-mail: yub@im.ac.cn.
Abstract: The regulation of the expression of genes involved in metabolic pathways, termed as metabolic regulation, is vital to construct efficient microbial cell factories. With the continuous breakthroughs in synthetic biology, the mining and artificial design of high-quality regulatory elements have substantially improved our ability to modify and regulate cellular metabolic networks and its activities. The research on metabolic regulation has also evolved from the static regulation of single genes to the intelligent and precise dynamic regulation at the systems level. This review briefly summarizes the advances of metabolic regulation technologies in the past 30 years.
Keywords: metabolic engineeringregulation of gene expressionsynthetic biologymicrobial cell factories
代谢工程的目标是通过改变和调控宿主体内的关键代谢途径或引入新的代谢途径来提高所需化学品和蛋白的产量等,包括关键酶催化底物范围的扩大、新型化学品的生产、产量的提高和细胞生产力与鲁棒性的增强等,涉及一系列不同的策略。在过去的30年里,代谢工程已经成为改善细胞性能、实现化合物可持续生物制造生产的一个有力工具,但代谢通路的改变及外源蛋白质的引入增加了对细胞资源的需求,打破了细胞正常的生理平衡。通路蛋白的过度表达会阻碍细胞生长,进而降低生产力;而简单降低途径酶的表达水平会形成反应途径瓶颈,从而限制了产率和效价。因此,需要对通路进行精细的调控,实现细胞生长和产物合成的平衡。代谢途径的优化一般是在转录水平和翻译水平上进行优化,包括启动子、RNA调控元件、蛋白质水平等的优化。优化代谢路径的策略包括静态调控和动态调控。静态调控包括调节启动子强度[1]、核糖体结合位点[2]或载体拷贝数[3],以实现路径反应通量平衡和消除瓶颈,从而提高产品产量。尽管静态调控的策略相当稳健,但菌株生长往往受到一定削弱,其产物生产速率也会降低。另外,若涉及细胞生长必需基因,对其进行静态调控有相当大的难度,甚至导致细胞死亡。因此,更先进的优化策略涉及蛋白质按需表达的动态调控。使用可诱导启动子的动态调控使生长期与生产期分离,允许细胞在引导资源形成所需化合物之前先积累足够的生物量[4-5]。此外,当宿主或环境条件发生变化时,通过感知关键中间产物来调节蛋白质表达水平的动态调控可使细胞实时调整其代谢流量[6-7]。这有助于调节细胞资源的有效消耗,保持最佳的蛋白表达水平用于合成目标产品。近年来,合成生物学的发展使得研究者可以通过更为智能化的方法对细胞表型实现时序或者空间水平上的调控。对细胞调控的改造技术也向着可编程、多靶点及自动反馈控制方向发展。
1 启动子工程启动子是具有RNA聚合酶识别、结合和转录起始功能的一段特殊的非编码DNA序列,其结构、稳定性以及与RNA聚合酶的亲和力在很大程度上会影响基因表达[8]。转录是基因表达过程的第一步,在启动子水平上调控特定基因的转录是调节代谢通量最直接、最有效的方式之一[1]。因此,代谢工程应用长期以来一直依赖于有效启动子的发现和表征。启动子工程包括对天然启动子的挖掘、生物功能特性表征及人工改造,是代谢工程和合成生物学研究中调节基因表达和优化代谢物生物合成的重要基础。代谢途径中对基因的微调表达是调控细胞代谢通量的关键步骤[9-11]。合理和精确地调控多基因途径中的基因表达可以显著影响代谢通量分布并使特定代谢产物的产生最大化[12]。基因表达水平必须与胞内和胞外条件的变化相协调,才能维持代谢网络的稳态,因此,动态调控基因表达需要更灵活的调控启动子[13]。目前已经进行了许多研究,包括挖掘天然启动子、构建融合型启动子、设计人工启动子在转录水平上微调特定基因的表达并提高目标代谢物的生产水平[14]。在本节中,我们将综述天然启动子的挖掘和改造,同时也概述了静态和动态启动子调控在代谢工程中的应用。
1.1 天然内源启动子挖掘微生物基因组中包含多种内源启动子,是启动子工程和代谢途径工程的巨大宝库[15]。迄今为止,许多内源启动子已被用于宿主中多种酶的过表达。Zaslaver等[16]利用绿色荧光蛋白GFP评价了大肠杆菌K12中2 000个启动子的强度。Zhou等[17]根据RNA-seq数据筛选并鉴定了104个来自大肠杆菌的天然启动子5′-UTR复合物,在转录水平上强度为PBAD启动子的0.007%至4 630%,以用作对大肠杆菌细胞中的转录网络进行准确分析的工具。枯草芽孢杆菌作为革兰氏阳性模式菌株已广泛用于合成生物学研究、代谢工程改造的宿主和工业酶的生产菌株,Song等[18]用GFP来评估枯草芽孢杆菌内源启动子的强度,总共测试了84个位于不同类别蛋白质上游的预测启动子序列,测得的表达水平是表征良好的强启动子P43活性的0.002 3倍至4.53倍,从中选择的启动子将胞质蛋白β-半乳糖苷酶和分泌蛋白α-淀粉酶的表达量分别提高了4倍和2倍。Yang等[19]筛选了枯草芽孢杆菌114个在生长不同阶段发挥活力的启动子,建立了可调控基因在不同生长阶段表达的启动子库。Zhu等[20]在酿酒酵母体系中建立了一个特异性启动子数据库,包含580个转录因子结合位点和425个转录起始位点。Keren等[15]对酿酒酵母中约900个启动子进行了分类和表征,这些天然启动子的发现将有助于体内的代谢途径调控改造。
1.2 启动子的人工改造天然启动子的局限性在于它们无法获得完整连续的转录调控强度,并且无法使生物体内的转录水平最大化。有限数量的可用内源启动子通常是微调转录调控的瓶颈[21-22]。通过对已有启动子进行突变改造,在大肠杆菌[12, 23-24]、谷氨酸棒杆菌[25]、酵母[12, 26-28]中可获得各种强度的工程启动子库。Liu等[29]在枯草芽孢杆菌168中创建了从头合成启动子文库,并用于肌苷和乙醛的生产,其产量分别增加了700%和44%。Li等[30]在盐单胞菌TD01中,对高表达蛋白Porin编码基因的上游进行一系列分子生物功能表征从而鉴定出启动子的核心区,在其–10区和–35区之间的序列进行随机突变得到一个强度范围为310倍的启动子库,并用于调控盐单胞菌TD01聚3-羟基丁酸酯的合成。
借助对大量启动子样本的分析和特定位点突变,可以总结出启动子序列的特征,将所需功能的各个结构部分连接在一起,可得到具有多种功能的新启动子。杂合启动子的构建是启动子工程的另一种有效策略。Jiao等[31]基于启动子PgroE和操纵子lacO融合构建了异丙基-β-d-硫代半乳糖苷(Isopropyl-β-d-thiogalactopyranoside,IPTG)诱导人工启动子Pg2,用于替换内源启动子PsrfA,在枯草芽孢杆菌中使表面活性素产量提高至5.98 g/L。通过将启动子PaprN的–10区替换为σA依赖性启动子的共有序列,使枯草芽孢杆菌中纳豆激酶活性提高136%[32]。在酵母菌中,重组已知的功能区域来创建新的杂合启动子同样可产生高表达、可调整强度的启动子文库[33-34],并实现对基因表达的微调,有效提高目的产物的产量[35-36]。例如,Madzak等[36]在解脂耶氏酵母中基于天然启动子PXPR2构建了一系列杂合启动子,使其表现出很强的组成型活性,不受碳源氮源或酸性条件的抑制,扩大了该强启动子在工业应用上的使用范围。
双启动子表达系统,定义为两个启动子组合使用来控制同一基因转录[37],是一种有效提高重组蛋白表达水平的方法[38-43],在工业应用中具有较大潜力。尽管双启动子的调节机制尚不清楚[43],但重组蛋白产量的提高表明该启动子工程策略是增强启动子活性的有效途径。例如,Zhang等[38]构建了双启动子PHpaII–PamyQ’系统,使枯草芽孢杆菌β-环糊精糖基转移酶的酶活力达到571.2 U/mL,证明了该系统在工业应用中的潜力。
当启动子的结构和作用与位点选择性诱变明确相关时,还可以使用数学计算和计算机模拟方法来预测各种启动子对表达水平的影响。基于能量矩阵,研究人员构建了一个合成的大肠杆菌启动子文库,包含20个经过理性设计的启动子,其强度为原始trc启动子的0.8%至100%[44]。Brewster等[45]通过使用序列依赖性分析结合计算模型与转录热力学模型,针对大肠杆菌RNA聚合酶σ70全酶设计了18个独特的转录因子结合位点,用以调节靶向基因的表达水平。
1.3 启动子对基因表达的静态调控通过改变启动子的强度来上调或下调基因的表达水平从而改变代谢通量,是提高目标物质产量的基本静态调节策略[14]。自代谢工程兴起以来,研究者们采用最多的菌株改造策略是静态调控。静态调控通过启动子替换常量表达外源酶,改造得到的菌株其表型在发酵过程中是单一的,产物的合成和生长过程是相耦合的。
1.3.1 单基因或基因簇的静态调控过强的表达会对菌体造成负担,启动子对基因表达的合理调控,对于工程微生物高效生产目标化学品至关重要。适当的基因表达水平的启动子可以在转录水平上提供强大的开关,用以微调单个基因或基因簇,这对于重建途径至关重要。Xu等[46]研究了不同启动子在大肠杆菌中对2, 3-丁二醇合成天然基因簇budABC表达的影响,结果表明,启动子Pabc的应用效果更好,最终使2, 3-丁二醇的产量达到21.5 g/L。Sun等[47]鉴定了酿酒酵母中14个内源性启动子,分别取代了玉米黄质生物合成的5个关键基因的内源启动子,所得菌株直接利用桦木木聚糖合成玉米黄质,产量为(0.74±0.02) mg/L。另有许多启动子被用于大肠杆菌[48-51]、枯草芽孢杆菌[52]、谷氨酸棒杆菌[53]等宿主中多种酶的过表达,这些酶参与了有工业价值代谢物生产的代谢途径,如生产胡萝卜素、2-吡喃酮-4, 6-二羧酸和聚乳酸等。
1.3.2 模块化的静态调控对于合成步骤较多的代谢途径改造通常涉及调控多基因的共表达,根据代谢合成靶基因的功能或中间物的特性,可将合成途径分成不同的模块进行分别优化。采用模块化策略组合调节各模块的协同表达,可以有效平衡途径(图 1A)。Shukal等[54]将甲羟戊酸途径基因分为两个代谢模块,优化了工程大肠杆菌的转录水平,实现了类异戊二烯的高产。Zhou等[55]将葡萄糖合成丹参素的途径划分为3个代谢模块进行染色体整合,最终在大肠杆菌中实现了5.6 g/L的高产。Wang等[56]将5-羟基色氨酸的生物合成途径分为3个功能模块,包括l-色氨酸模块、羟基化模块和辅因子供给模块,发现降低辅因子模块的启动子强度,可达到途径的较优平衡,实现目前报道的微生物产生5-羟基色氨酸的最高记录。黄酮类化合物的发酵途径通常涉及多个外源基因的组装,启动子工程模块化被广泛应用。通过修饰质粒基因拷贝数和启动子强度,实现了松属素[57]、白藜芦醇[58]和柚皮素[59]等的高水平合成。
 |
| 图 1 基于不同启动子调控基因表达的原理示意图 Fig. 1 Schematic diagram of the principles of regulating gene expression by using different promoters. (A) Static modularization control. (B) Inducer regulation. (C) Metabolite concentration regulation. (D) Quorum sensing regulation. |
| 图选项 |
1.4 启动子动态调控通过静态调控策略,产物合成途径的代谢流可以得到有效加强。然而代谢是广泛重要且复杂的细胞过程,依赖于静态调节基因表达难以满足对目标化学品生产代谢网络中多基因协同表达的有效精确调节。尤其当产物合成途径是利用菌株原本用于生物质合成的代谢产物作为前体时,静态调控往往达成的是一个菌株在目标产物生产和细胞生长之间的被动平衡状态,即产物转化率的提高某种程度上是以牺牲细胞生长为代价的。相反,动态调控可以控制蛋白质按需表达,对于生物催化剂的工程设计和优化是有效的,它可以使细胞生长良好,合成能力提高。动态调节网络的构建与微生物的自然调节机制非常相似,通过感知环境和细胞内信号来实时调节细胞的代谢状态,已成为代谢途径工程的一种有前景的策略[60]。在过去的几十年中,已经开发了很多基于外界环境条件诱导、基于代谢物浓度传感器或依赖于细胞密度的群体感应自主诱导的动态调控系统,通过平衡细胞生长和产物合成来实现产量最大化。
1.4.1 基于外界条件的诱导调控诱导性启动子可对代谢途径进行动态调控,其提供了一种分离生长阶段和产物合成阶段的方法。在添加诱导剂之前,细胞仅为正常生长模式,用于产物合成相关基因不能表达。当细胞浓度达到所需的阈值时,添加诱导剂激活相关酶的表达,可避免代谢产物的流失(图 1B)。Nakashima等[61]构建了基于木糖诱导的表达系统,成功地利用葡萄糖和木糖在大肠杆菌中生产了54 g/L的2, 3-丁二醇和31 g/L的乙酰丁酸。Kim等[62]利用Aro80半结合位点和Aro9核心启动子元件的重复序列构成芳香族氨基酸诱导型启动子,用其表达alsS和alsD基因来生产乙偶姻,从而提高酿酒酵母的乙偶姻产量。除了最常用的IPTG、脱水四环素诱导型启动子之外[63],其他发酵过程条件控制诱导型启动子已广泛用于代谢调控,如温度、pH、光照等。其中温度调控在发酵控制中操作相对简单,已经得到广泛的应用,例如Zhou等[64]利用噬菌体λ来源的PR和PL融合启动子替换d-乳酸脱氢酶的ldhA启动子,构建了热诱导启动子,在42 ℃实现生产模式与生长模式的转换,从而提高了d-乳酸的产量。pH值也可作为触发条件来实施动态途径工程。利用系统工程改造的启动子PYGP1和PCCW14,在酵母中实现了低pH条件下启动基因表达来调控乳酸的生产[65]。在另一个实例中,PGAS被鉴定为pH响应性启动子,以调控顺式-乌头酸脱羧酶(由cad编码) 的表达,转录阻遏物在低pH下失活,以诱导cad表达并提高衣康酸的产量[66]。通过构建光遗传学开关以光照调控基因表达水平,也是一种有吸引力的调控手段,例如,Senoo等[67]使用CcaS/CcaR系统,在绿光照射下实现了大肠杆菌中糖酵解和甲基乙二醛途径之间光诱导的代谢通量调控。
1.4.2 基于代谢物浓度的动态调控代谢物响应性启动子通常来自天然启动子,其首先通过用所需代谢物进行筛选和鉴定,然后通过操纵启动子强度来优化动态范围(图 1C)。使用这样的启动子的一个优点是它仅涉及启动子替换,而无需表达额外的转录因子[68]。例如Yuan等[69]在酵母中筛选了天然麦角固醇响应性启动子,并用于下调ERG9的表达,将中间物的代谢通量转移至紫穗槐二烯,使产量提高了5倍。
另一种代谢物响应调控系统涉及生物传感器(Biosensor),通常源自与代谢物相互作用后结合DNA的转录因子。根据宿主的不同,需要对转录因子和启动子上的结合位点进行人工改造才能获得所需的动态范围[70-71]。Xu等[72]改变PdhR在启动子上的结合序列与位置,优化了丙酮酸响应基因回路的动态范围,实施了有效的动态调控,提高了枯草芽孢杆菌的葡萄糖二酸产量。基于FapR/FadR设计的生物传感器,已有多篇报道用于研究辅酶A与细胞生长之间的平衡代谢[7, 73-74],提高大肠杆菌中脂肪酸合成的效率。Xu等[73]通过整合转录因子FapR和PGAP的顺式作用模块(Cis-regulatory module) 构建了丙二酰辅酶A传感器,通过该生物传感器使丙二酰辅酶A与细胞生长之间达成代谢平衡,脂肪酸产量显著提高。这种FapR/丙二酰辅酶A生物传感器也已被运用到酵母中,动态诱导异源丙二酰辅酶A还原酶的表达以产生3-羟基丙酸[75]。另外,大肠杆菌转录因子调节子PcaU可被3, 4二羟基苯甲酸诱导[76],利用该调节子构建了对羟基肉桂酸有响应的生物传感器,并可用于提高香兰素和对羟基苯甲酸酯的产量[77]。
除了天然存在的调节代谢,人工构建调节组件来动态调控多层代谢网络具有更广泛的应用价值。Chen等[78]对大肠杆菌调节蛋白AraC进行了工程设计,使其可以识别四氢嘧啶为非天然效应物,开发了高通量筛选系统,进一步提高了四氢嘧啶生物合成簇的生物合成效率。Liang等[79]设计出了尿酸响应调节蛋白HucR,其对产物香兰素和底物阿魏酸都有不同的反应能力,通过早期生长阶段产物途径的抑制及后期产物反馈激活通路表达显著提高了生物合成效率。
1.4.3 基于群体感应的调控系统群体感应(Quorum sensing,QS) 是细菌细胞间通讯的一种形式[80]。群体感应系统提供了另一种自动感应的方法,即当某种细胞团积聚时,该方法就会触发。表 1列举了目前研究较多的群感效应分子。
表 1 目前研究较多的几种细胞间通讯系统(数字代表对应的菌株来源)Table 1 Summary of the reported intercellular communication systems. The numbers in brackets shown in the superscript in the column of "Synthase" and "Receptor" correspond to the numbers shown in the column of "Source", indicating the source of a particular gene.
| Inducer | Synthase | Receptor | Promoter | Source | References |
| Isovaleryl | BjaI | BjaR | Pbja | Bradyrhizobium japonicum | [81] |
| p-coumaroyl | RpaI | RpaR | Prpa | Rhodopseudomonas palustris | [82] |
| Salicylate | pchBA(2) | NahR(1) | Psal | (1) Pseudomonas putida (2) Pseudomonas aeruginosa (3) Yersinia enterocolitica | [83] |
| Irp9(3) | |||||
| 2, 4-diacetylphloroglucinol | PhlACBD | PhlF | PphlF | Pseudomonas fluorescens | [84] |
| Methylenomycin furan | MmfLHP | MmfR | PmmfR5 | Streptomyces coelicolor | [85] |
| Naringenin | CHS(2), CHI(3) | FdeR(1) | PfdeA | (1) Herbaspirillum seropedicae (2) Petunia X hybrid (3) Medicago sativa | [86] |
| C4 | RhlI | RhlR | Prhl | Pseudomonas aeruginosa | [87] |
| C6 | LuxI | LuxR | Plux | Vibrio fischeri | [88] |
| C8 | CepI | CepR | PcepI | Burkholderia cenocepacia | [89] |
表选项
以来自费氏弧菌的LuxR和产生酰基高丝氨酸内酯(Acyl homoserine lactone,AHL) 的LuxI酶组合为例,AHL与LuxR转录因子相互作用,形成可以激活基因表达的LuxR/AHL复合物[88]。由于AHL的浓度与细胞浓度成正比,达到阈值时,将生成LuxR/AHL复合物并激活基因表达,可以在AHL积累后表达细胞密度依赖性基因。通过调控luxI和luxR的表达水平,可改变触发基因表达的切换时间(图 1D)。利用LuxR系统进行动态途径调控,在不添加外部诱导剂的情况下,可使得大肠杆菌合成没药烯的产量达到1.1 g/L[90]、高效合成1, 4-丁二醇[91]以及通过触发切换开关将三羧酸循环通量重定向为异丙醇的生产[92]。
在酵母系统中,因为真核生物转录抑制的特性,上述原核生物常用的群体感应系统在代谢工程改造的应用受到限制。Williams等[93]设计出响应细胞外α型信息素的方式来实现细胞间通信,成功实现种群密度依赖的GFP蛋白的表达。基于此设计出高种群密度自主触发基因表达,与RNA干扰模块连接以实现靶基因沉默的基因线路,实现对莽草酸途径通量的调控,用于对羟基苯甲酸的合成[94]。
2 RNA调控元件用于代谢调控改造由于RNA在感知分子配体方面的多功能性,而且与蛋白质相比,RNA元件可以更容易地建模和进行设计,越来越多的研究专注于利用RNA对代谢途径中的基因进行表达调控[95]。目前报道的RNA元件用于代谢调控的改造主要包括小分子RNA、核糖开关、RNA干扰和反义RNA技术。
2.1 合成小分子调控RNA基因敲除实验通常是功能基因组学和代谢工程研究中必不可少的实验过程。然而,重复的多个基因敲除费力费时,并且对于细胞功能至关重要的基因是不可行的。这就需要开发非敲除的分子工具来实现代谢途径调控。小分子RNA (sRNA)是原核生物中的非编码短RNA,可以有效地在翻译水平调控靶基因的表达,实现代谢过程的调控(图 2A)[96]。在琥珀酸酯的生产过程中,前体磷酸烯醇丙酮酸(Phosphoenolpyruvate,PEP) 被消耗用于葡萄糖的运输和磷酸化,并且产生大量的副产物,这是阻止琥珀酸盐生产提高的关键障碍。Zhao等[97]选择PEP作为靶标,通过引入抗pykF的合成sRNA,最终优化菌株中乳酸和乙酸盐含量明显降低,琥珀酸滴度为30.12 mmol/L。代谢通量分析和发酵动力学表明,该方法可以减轻细胞ATP供应的限制和氧化还原的不平衡,进而改善大肠杆菌中的琥珀酸盐的生产。Yang等[98]开发了在大肠杆菌中用于快速、多重基因组规模靶基因敲低的合成sRNA表达平台。基于该技术平台,迅速开发出l-脯氨酸(54.1 g/L) 和l-苏氨酸(22.9 g/L) 的高性能菌株。此外,在1 858个合成sRNA文库中,利用高通量筛选技术快速找到靶向基因,获得紫丁香素(5.19 g/L) 和靛蓝(135 mg/L) 的高产菌株。基于此sRNA文库,作者还找到14个敲低后能提高大肠杆菌中丙二酰辅酶A水平的基因。这些靶向基因被应用于提高聚酮化合物(6-甲基水杨酸和芦荟酮) 和苯丙类化合物(白藜芦醇和柚皮素) 的产量,产量分别达到440.3、30.9、51.8和103.8 mg/L[99]。
 |
| 图 2 四种常见的RNA调控机制的原理示意图 Fig. 2 Schematic view of different RNA regulatory mechanisms. (A) Synthetic regulatory small RNA. (B) Riboswitch. (C) CRISPRi. (D) asRNA. |
| 图选项 |
为了调节sRNA对靶标mRNA的作用强度,可使用不同启动子调节合成sRNA的丰度,从而实现对基因表达敲低系统的精细调控[100]。Noh等[101]选择热通量响应计算分析和与产品生物合成相关的靶基因,通过sRNA-启动子组合文库转化菌株的筛选来提高目标产物的产量。利用这种方法,对argF和glnA进行微调,降低该基因表达水平,通过补料分批培养分别实现腐胺((42.3±1.0) g/L)和l-脯氨酸((33.8±1.6) g/L) 的高水平合成。Sung等[102]通过改造启动子的强度,调节靶向基因murE的sRNA胞内水平,构建的工程菌株可产生2.15 g/L尸胺。谷氨酸棒杆菌也是重要的平台菌株,广泛用于氨基酸和各种其他生物化学物质的工业生产中。Sun等[103]针对谷氨酸棒杆菌开发了基于合成sRNA的基因敲低策略,降低靶向基因pyk、ldhA和odhA的表达,使谷氨酸滴度和产量提高了近3倍,相关基因的转录和酶活性下调了80%。
2.2 核糖开关核糖开关(Riboswitch) 可以感知各种生化和物理信号,大幅度提高对复杂细胞行为进行编程的能力。核糖开关通过特异性结合代谢物,可对mRNA分子5′非翻译区进行调控表达,使RNA结构重排,从而调控转录的翻译起始或过早终止,是调节工业微生物中代谢通量调控的有效工具(图 2B)[104]。
多数成功设计的核糖开关在某种程度上依赖于高通量筛选和/或从基因开关的组合文库中进行选择。有报道通过筛选核糖开关的通用高效文库,得到新型硫胺素焦磷酸核糖的人工开关,该人工核糖开关的活性提高了58倍[105]。此外,也有报道称成功筛选获得了对抗生素新霉素有响应的合成转录核糖开关[106]。Boumezbeur等[107]开发了一种新的理性改造核酸开关的方法:通过删除一段关键核苷酸序列,可有效防止终止发夹的形成,将核酸开关恒定地保持在打开状态,特定序列的删除不会影响其他二级和三级结构的形成,不会影响基因的其他功能。通过改造鸟嘌呤感应核糖开关(pur) 和黄素单核苷酸感应核糖开关(rib),使关键基因表达水平比对照提高了90倍,工业核黄素高产菌的生长得到有效改善(生物量增加了53%),且核黄素产量有大幅提升。
细菌体内的天然核糖开关还可作为氨基酸生产的感应元件。比如,大肠杆菌和枯草芽孢杆菌中存在天然的赖氨酸核糖开关,它们均可控制柠檬酸合酶的表达,从而调控三羧酸循环中的代谢通量[108]。Zhou等[109]将大肠杆菌来源的天然“赖氨酸-关闭”核糖开关改造成“赖氨酸-打开”核糖开关,并用来调控赖氨酸的合成。将选定的“赖氨酸-打开”核糖开关导入谷氨酸棒杆菌中,用于调控lysE基因(编码赖氨酸转运蛋白) 的表达,在重组谷氨酸棒杆菌中实现赖氨酸转运的动态调控,赖氨酸产量比出发菌株提高了21%。
2.3 CRISPR干扰CRISPR干扰(CRISPR inference,CRISPRi)使用核酸酶活性失效的Cas9蛋白,在gRNA的引导下结合到序列碱基互补配对的靶基因上,从而干扰基因的正常表达(图 2C)。CRISPRi的机制及其在微生物生理和进化中有重要作用,这种新型干扰技术已经在细菌中表现出了潜在的应用价值[110]。尽管仍处于起步阶段,CRISPRi在大肠杆菌、枯草芽孢杆菌、谷氨酸棒杆菌等代谢工程菌株中已有较多的应用(表 2)。本节将重点介绍CRISPRi在这3个常用宿主系统中的应用实例。
表 2 CRISPRi技术用于代谢工程调控改造的实例总结Table 2 Summary of the applications of CRISPRi in metabolic regulation
| Organisms | Biotechnological applications | Gene targets | References |
| E. coli | Polyhydroxybutyrate | phaC | [118] |
| E. coli | Polyhydroxybutyrate | dxs, glmU, murA, murC, murD, murE, mraY, ftsW | [119] |
| E. coli | Polyhydroxybutyrate | ftsZ, mreB | [120] |
| E. coli | Polyhydroxybutyrate | gltA | [121] |
| E. coli | 1, 4-butanediol | gabD, ybgC, tesB | [122] |
| E. coli | n-butanol | pta, frdA, ldhA, adhE | [123] |
| E. coli | 5-aminolevulinic acid | hemB | [124] |
| E. coli | Malate | acn, ms | [125] |
| E. coli | Pinosylvin | fabD | [126] |
| E. coli | Pinocembrin | metK, proB | [127] |
| E. coli | Arginine | argR | [128] |
| B. subtilis | Surfactin | yrpC, racE, murC | [129] |
| B. subtilis | Amylase | epr, nprB, mpr, bpr, vpr, wprA | [130] |
| B. subtilis | N-acetylglucosamine | zwf, pfkA, glmM | [131] |
| B. licheniformis | l-valine | alsD, bcd | [132] |
| C. glutamicum | l-lysine | pyc, gltA, idsA, glgC | [133] |
| C. glutamicum | Shikimic acid | pyk, ncgl2809, sugR | [134] |
| C. glutamicum | Citrulline | argH, pgi | [135] |
| C. glutamicum | Putrescine production | carB, ilvH, ilvB, aroE | [136] |
| S. coelicolor | Actinorhodin | actinorhodin pathway genes | [137] |
| S. pneumoniae | Teichoic acid | murT, gatD, SPD_1198(tarP), SPD_1197(tarQ) | [138] |
| P. putida | Pyoverdine synthesis | pvdH | [139] |
表选项
2015年,Lv等[111]首次报道用CRISPRi调控中心代谢途径的基因表达,用于调节PHA合成中4-羟基丁酸组分的比例。乌头酸是三羧酸循环中的一个重要的中间代谢产物,Li等[112]在大肠杆菌中利用CRISPRi系统同时靶向丙酮酸激酶和异柠檬酸脱氢酶。在摇瓶和分批补料培养中,该CRISPRi菌株产生的乌头酸相对于对照菌株分别提高了60倍和15倍,并且该菌株的乙酸和乳酸等副产物一直维持在低水平,说明CRISPRi系统可以通过协调糖酵解和三羧酸循环来有效改善乌头酸的生产。CRISPRi在代谢工程领域的应用还包括平衡同一代谢通路中上下游基因表达的平衡。例如,Kim等[113]利用CRISPRi技术,在大肠杆菌中优化甲羟戊酸途径基因表达实现了(-)-α-红没药醇和番茄红素的增产,并减轻了多基因表达或途径有毒中间代谢物积累对细胞生长的抑制作用。
利用CRISPRi还可抑制细胞生长必需基因的表达,在合适的时机停滞细胞生长,以提高产物生产的转化率等。Cleto等[114]在谷氨酸棒杆菌中CRISPRi系统使用失活的Cas9 (dCas9) 来抑制靶向基因。该研究将pgi和pck基因的表达降低了98%,将pyk基因的表达降低了97%,产生的l-赖氨酸和l-谷氨酸的滴度提高比率可与基因删除所达到的水平一致。Westbrook等[115]在生产透明质酸的枯草芽孢杆菌中应用CRISPRi来分别减少出发菌株中pfkA或zwf基因的表达,从而显著提高了透明质酸的合成水平。
由于CRISPRi/dCas9系统在多靶点智能调控的应用场景中还有一些限制,包括遗传稳定性较差、对细胞有较大的代谢负载等。与之相比,对于多靶点调控,CRISPRi/dCpf1在应用上更为简便。dCpf1自身就具有RNA酶活性,可将多gRNA串联的pre-crRNA加工成分离的gRNA发挥作用。一些研究者也已尝试使用CRISPRi/dCpf1为基础构建的动态开关系统。Zhang等[116]使用CRISPRi/ dCpf1在耶氏解脂酵母Yarrowia lipolytica中实现了调控外源脱氧紫色杆菌酸途径基因vioA、vioB和vioE的表达量。Ji等[117]使用CRISPRi/dCpf1调节大肠杆菌生长必需基因的表达时机,提高了丁烯酸的合成效率。
以上报道表明,CRISPRi是系统调节生物合成和内源基因表达的有效工具,可用于调节生物合成代谢途径,尤其对处于代谢中心位置的化学品或中间物具有生物毒性的化学品的合成具有重要价值。它的应用可以促进微生物“智能细胞”工厂的技术发展,用来生产更多有工业价值的产品。
2.4 反义 RNA (asRNA)反义RNA (Antisense RNA,asRNA) 是一种研究比较深入的RNA调控元件。asRNA调节已用于许多代谢工程研究中,以优化靶基因的表达水平。asRNA也是研究必需基因敲低的有效工具[140-143]。大肠杆菌作为重要的外源蛋白表达宿主,生长过程中产生的乙酸会抑制蛋白表达。Kim等[144]采用asRNA策略降低了大肠杆菌的乙酸合成水平,将目标蛋白的总量提高了2.1倍。asRNA技术也可用于动态调节内源代谢过程来平衡细胞生长和途径合成效率的要求。例如,利用asRNA系统调控葡萄糖激酶的表达量,从而调控糖酵解通量,在不改变最终生物量积累的情况下,将工程化大肠杆菌的比增长率降低了50%[145]。将asRNA作用于代谢途径中靶标基因,可以改善最终产物的产量。Kang等[146]将asRNA应用于琥珀酸合成菌株的改造,产量提高了7倍。利用asRNA技术,Meng等[147]将大肠杆菌6-脱氧赤藓醇B的合成水平提高了296.2%。Yang等[148]改善了大肠杆菌胞内丙二酰辅酶A的含量,使4-羟基香豆素、白藜芦醇和柚皮素的产量分别提高了2.53倍、1.70倍和1.53倍。
asRNA技术在革兰氏阳性菌中也得到了广泛的应用。Wang等[149]利用asRNA调控了枯草芽孢杆菌中生物素羧化酶Ⅱ的表达基因yngH的表达,使表面活性肽产量提高到13.37 g/L,比对照提高了43%。Rebekka等[150]采用asRNA策略沉默关键基因hemZ,减少巨大芽孢杆菌四吡咯途径的血红素分支的通量,显著提高了细胞内维生素B12的浓度。Desai等[151]将丙酮丁醇梭菌中丁酸激酶和磷酸反丁酰化酶作为靶点,两个酶的活性分别比对照低70%和80%,工程菌的生长量比对照低28%,该实验还表明丁酸酯合成通量不受丁酸酯合成酶的水平调控。Tummala等[152]研究了asRNA结构性质对丙酮丁醇梭菌ATCC 824中丙酮形成途径关键酶——乙酰乙酸脱羧酶和辅酶A转移酶下调功效的影响,证实辅酶A转移酶是丙酮合成过程的限速酶。
3 翻译水平调控翻译的调节主要发生在起始阶段,该过程主要涉及核糖体和RBS的结合,RBS的识别和结合受到温度、小分子化合物、蛋白质或反义RNA的结合调节,从而影响基因的表达水平。另一方面,密码子使用的偏好性,即对某些同义密码子的偏好是所有基因组的普遍特征。越来越多的证据表明密码子使用频率通过影响共翻译蛋白质折叠、翻译效率和准确性、mRNA稳定性和转录可以调节蛋白质结构和基因表达。
3.1 核糖体结合位点(RBS)工程在翻译起始阶段被核糖体覆盖的mRNA片段被称为核糖体结合位点(RBS)[153]。在细菌中,RBS通过互补配对与16S rRNA的3′末端序列相互作用。参与此相互作用的碱基对的数量以及这些碱基对相对于起始密码子的位置决定了翻译效率(图 3A)[154]。进一步研究认为,通过二级结构的形成来增强RBS区域的稳定性会对翻译效率产生负面影响。对RBS二级结构如何决定翻译效率的定量分析结果表明,仅暴露RBS区域或起始密码子都不会被核糖体优先识别,核糖体也不能识别RBS区域和起始密码子之外的核苷酸,翻译效率与未折叠RBS的mRNA分子的比例密切相关[155]。
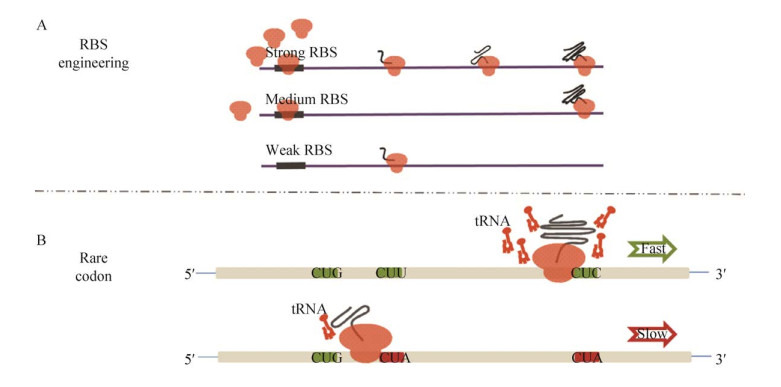 |
| 图 3 两种翻译水平调控机制的原理示意图 Fig. 3 Schematic view of two translation-level regulatory mechanisms. (A) RBS engineering. (B) Gene translation regulated by rare codons. |
| 图选项 |
核糖体结合位点计算器(RBS calculator) 是一种用于预测和调控细菌翻译起始和蛋白质表达的设计方法。该方法可以预测mRNA转录本中每个起始密码子的翻译起始率,还可以优化合成RBS序列以获得目标翻译起始率。利用RBS计算器,蛋白质编码序列的翻译率可以在0.001到10万倍的比例范围内得到合理的调控。因此,其合理调控蛋白表达水平的能力在生物技术中有着广泛的应用。近年来,根据mRNA转录物和30S核糖体复合物之间的分子相互作用强度来量化翻译起始速率,开发了人工合成RBS的自动化设计工具[2, 156-158],利用RBS计算器可设计相应的酶表达水平,从而部分消除合成途径的瓶颈[4, 158-161]。人工RBS策略已经用于提高脂肪酸、异戊二烯、丙二醇、d-对羟基苯甘氨酸等生物合成[160, 162-164]。未来RBS计算器还能用来优化基因线路,合理调整基因回路中蛋白质的表达水平。通过RBS计算器将基因线路的定量模型转换成一个合成的RBS序列,可选择性地增加或降低蛋白质的表达水平。
3.2 稀有密码子调控蛋白表达翻译效率以及蛋白质生产效率,是由翻译起始速率和延伸速率决定的。翻译起始速率控制着转录产物被翻译的频率,而翻译延伸速率控制着翻译过程的速度。翻译起始阶段,核糖体与mRNA结合,并且起始密码子必须被识别。起始密码子只包含3个核苷酸,但它在翻译水平上显著影响基因表达强度[165]。ATG是最常见的起始密码子,但一些基因也使用GTG,而TTG则更为罕见[166-167]。还有研究发现GTG的翻译起始效率低于ATG,有时会用ATG代替GTG以增加靶基因的表达[168]。Wu等[169]以RFP、GFP和lacZ作为报告基因,在大肠杆菌中构建了动态范围大、表达水平均匀的起始密码子库,开发了一种利用起始密码子组合调控优化代谢途径的新策略,并将其应用于大肠杆菌玉米黄质合成途径3个基因crtZ、crtY和crtI的调控,最后筛选到一株玉米黄质产量为6.33 mg/L的菌株,产量比出发菌株提高了10倍。
在较高的翻译起始速率下,还需要较高的翻译延伸速率才能获得基因的最佳表达[170]。低起始速率或低翻译延伸速率都可独立引起蛋白的低表达。稀有密码子通常由于其同源tRNA的稀有性而阻碍翻译,因此,稀有密码子的这种特性可以用来调节蛋白质的表达(图 3B)。作为大肠杆菌中最稀有的密码子,AGG、AGA、CUA、AUA、CGA和CCC在调节不同内源性蛋白的表达中起重要作用[171-172]。这些稀有密码子在基因中的相对位置与大肠杆菌中蛋白质表达被抑制的水平有关[173-174],例如,Finger等[175]在大肠杆菌中表达类马链球菌Streptococcus equisimilis来源的链球菌激酶基因skc,并在+2、+3、+5、+8、+9和+11等不同位置选择性地引入了AGG密码子。结果表明,在+2、+9和+11位置引入“AGG”有助于蛋白过表达,而移到+3和+5位置则无表达。冯凡等[176]在构建异戊二烯合成菌株时,为了平衡整个代谢途径,需要降低途径关键酶乙酰辅酶A酰基转移酶/HMG-CoA合成酶的表达量,在基因中引入了2个AGG稀有密码子,同时组合优化其他关键酶的表达量,将异戊二烯产量提高了73%。
在翻译延伸率方面,一般认为使用被高丰度tRNAs识别的密码子,翻译延伸速度越快,翻译效率也越高[177]。因此,提高基因表达的一个常规策略就是替换目的基因中的稀有密码子,使它们与宿主的密码子偏好性更为匹配,而不改变被编码蛋白的氨基酸序列。以大肠杆菌[178-181]和酿酒酵母[182-183]为宿主的异源蛋白表达实验也证明了密码子优化在一定程度上能够提高蛋白表达强度。然而,近年来又有许多核糖体分析数据的研究表明,高翻译延伸率和常用密码子之间没有相关性[184]。一些真核生物的研究表明,常用密码子的翻译速度与稀有密码子的翻译速度相同[185-186]。最近的研究表明,同源tRNAs数量较少的稀有密码子译码速度较慢,从而导致翻译延伸率降低[187-188]。这些结果表明,密码子偏好性适应于胞内tRNA池,作为平衡tRNA供给和需求的策略,从而实现最佳的蛋白翻译。
4 翻译后调控在细菌基因功能和调节生物回路的研究中,已经开发了许多在转录水平和翻译水平上调控基因表达的方法。但这些方法也存在缺陷,即它们只能终止新的基因产物的合成,但无法消除预先存在的蛋白分子的影响。预先存在的产物主要通过细胞生长和分裂被稀释。对于长寿命蛋白,可能需要数个细胞周期才能将其细胞内数量和活性降低到不足以发挥作用的水平,在此期间,基因调控的时效性可能会受到很大影响[189]。蛋白质降解是调节细胞内信息的一种有效的天然机制,在维持细胞内稳态中起着至关重要的作用。通过失活或降解发挥功能的蛋白质浓度的方法也逐步用于代谢工程的动态调控研究中。蛋白质降解系统不仅能单独调控某一蛋白的特定降解,还能在合成生物学基础上开发可编辑的工具来执行复杂的功能,例如在工业微生物中利用蛋白质水平调控来实现代谢流的再分配[190-191];在生物过程中调控基因表达来实现实时的代谢控制并协调细胞反应[192-193];通过动态开关将细胞生长和产物合成解偶联从而提高微生物细胞工厂的工作效率[194]。
SsrA系统是细菌体内最重要的蛋白质浓度调控系统之一[195],此蛋白质降解系统发现于1996年,此系统的核心是通过蛋白酶对降解标签SsrA的特异性识别实现靶蛋白的快速降解[196]。近年来,已经开发出来一系列基于SsrA系统的新型基因调控工具,并显示出对合成生物学研究中生物电路设计调控的巨大潜力。McGinness等[197]在大肠杆菌中设计了一系列SsrA标签突变体,这些突变体减弱了与ClpXP蛋白酶的相互作用。在大肠杆菌中,衔接子SspB将SsrA标记的底物锚定在ClpXP蛋白酶上,可以用来调控目标蛋白的降解。当SspB衔接子存在时,ClpXP对被SsrA标记的靶蛋白的降解效率提高100倍。更重要的是,在体内不存在SspB衔接子时,带有这些标签的底物很稳定,仅在SspB诱导表达后,带有这些标签的底物才被迅速降解(图 4)。基于SsrA系统,在大肠杆菌中开发了一种动态调节宿主细胞内酶丰度的策略,并将其应用于葡萄糖生产肌醇的体系中[198-199]。通过调节糖酵解关键酶——磷酸果糖激酶-1在胞内的蛋白浓度,使肌醇的产量提高了2倍。Durante-Rodríguez等[200]基于此系统构建的FENIX系统,既能独立调控蛋白质的稳态水平,又能诱导靶蛋白的积累。利用该系统将细菌生长过程与PHB的积累解偶联。通过标记3-酮酰基-CoA硫脂酶(PhaA),催化合成路径的第一步反应,在乙酰辅酶A节点处建立了一个动态的代谢开关。当系统激活时,这种代谢前体可以有效地重新定向到PHB形成中。该工程菌在葡萄糖培养基以生长-生产非偶联模式获得了非常高的血红蛋白积累率(0.4 h?1),聚合物含量约为72% (W/W)。SsrA系统不仅可以在大肠杆菌中诱导蛋白降解,在革兰氏阳性菌中也已被成功应用。Griffith等[201]对大肠杆菌和新月柄杆菌Caulobacter crescentus来源的SsrA降解标签SspB衔接子进行改造,使其适应于枯草芽孢杆菌,同样实现了对靶蛋白的诱导降解。这也验证了两种不同来源的SsrA变体可在同一细胞内独立调控不同蛋白质的降解。
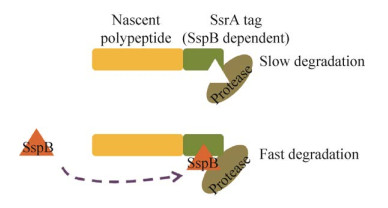 |
| 图 4 基于SsrA系统的蛋白降解调控机制的原理示意图 Fig. 4 Schematic view of protein degradation regulation by SsrA system. |
| 图选项 |
5 总结与展望代谢工程通过改造底盘细胞的代谢网络来提高细胞的合成能力,而合成生物学旨在利用机械工程原理创造新的和/或对现有细胞功能进行重新编程[202-205]。人工设计并合成的调控元件理论上与底盘细胞内源的转录调控网络相互作用较少,具有正交性[84]。因此,基于标准化表征元件构建可预测的、正交化的、定量可控的基因电路,有望实现细胞中基因“从头到尾” (Ground-up) 的均衡表达[206]。目前的研究结果也表明,利用合成生物学技术可以实现对代谢网络的多水平协同调控,显著提升了生物合成的能力[2, 207-213]。但需要强调的是,实现精准有效的平衡菌体生理和目标化合物的高效合成是工业微生物技术追求的极致目标。代谢工程对生产性能的经济性有严苛的标准,相比于一般的合成生物学研究,应用于实际代谢工程改造的调控工具对于元件的负载和毒性有着更为严格的限制[214]。尽管近年来合成生物学调控工具库被极大地充实,但是目前可用的开关工具相比于代谢工程多样的应用场景和不断升级的技术需求仍十分有限,需进一步优化并开发新的代谢工程应用导向的智能化动态调控系统。我们相信,随着合成生物学和人工智能技术的不断进步,将会有越来越多的表达元件与调控手段被开发,并应用于更广泛的生物制造过程。
参考文献
| [1] | Blazeck J, Alper HS. Promoter engineering: Recent advances in controlling trancription at the most fundamental level. Biotechnol J, 2013, 8(1): 46-58. DOI:10.1002/biot.201200120 |
| [2] | Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol, 2009, 27(10): 946-950. DOI:10.1038/nbt.1568 |
| [3] | Jones KL, Kim SW, Keasling JD. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab Eng, 2000, 2(4): 328-338. DOI:10.1006/mben.2000.0161 |
| [4] | Holtz WJ, Keasling JD. Engineering static and dynamic control of synthetic pathways. Cell, 2010, 140(1): 19-23. DOI:10.1016/j.cell.2009.12.029 |
| [5] | Lo TM, Teo WS, Ling H, et al. Microbial engineering strategies to improve cell viability for biochemical production. Biotechnol Adv, 2013, 31(6): 903-914. DOI:10.1016/j.biotechadv.2013.02.001 |
| [6] | Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol, 2000, 18(5): 533-537. DOI:10.1038/75398 |
| [7] | Zhang FZ, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol, 2012, 30(4): 354-359. DOI:10.1038/nbt.2149 |
| [8] | Solovyev VV, Shahmuradov IA, Salamov AA. Identification of promoter regions and regulatory sites//Ladunga I, Ed. Computational Biology of Transcription Factor Binding. Totowa, NJ: Humana Press, 2010, 674: 57-83. |
| [9] | Chen XL, Gao C, Guo L, et al. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem Rev, 2018, 118(1): 4-72. DOI:10.1021/acs.chemrev.6b00804 |
| [10] | Raman K, Chandra N. Flux balance analysis of biological systems: applications and challenges. Brief Bioinform, 2009, 10(4): 435-449. DOI:10.1093/bib/bbp011 |
| [11] | Troein C, Ahrén D, Krogh M, et al. Is transcriptional regulation of metabolic pathways an optimal strategy for fitness?. PLoS ONE, 2007, 2(9): e855. DOI:10.1371/journal.pone.0000855 |
| [12] | Alper H, Fischer C, Nevoigt E, et al. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA, 2005, 102(36): 12678-12683. DOI:10.1073/pnas.0504604102 |
| [13] | Zhou SH, Du GC, Kang Z, et al. The application of powerful promoters to enhance gene expression in industrial microorganisms. World J Microbiol Biotechnol, 2017, 33(2): 23. DOI:10.1007/s11274-016-2184-3 |
| [14] | Xu N, Wei L, Liu J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J Microbiol Biotechnol, 2019, 35(2): 33. DOI:10.1007/s11274-019-2606-0 |
| [15] | Keren L, Zackay O, Lotan-Pompan M, et al. Promoters maintain their relative activity levels under different growth conditions. Mol Syst Biol, 2013, 9: 701. DOI:10.1038/msb.2013.59 |
| [16] | Zaslaver A, Bren A, Ronen M, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods, 2006, 3(8): 623-628. DOI:10.1038/nmeth895 |
| [17] | Zhou SH, Ding RP, Chen J, et al. Obtaining a panel of cascade promoter-5?-UTR complexes in Escherichia coli. ACS Synth Biol, 2017, 6(6): 1065-1075. DOI:10.1021/acssynbio.7b00006 |
| [18] | Song YF, Nikoloff JM, Fu G, et al. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLoS ONE, 2016, 11(7): e0158447. DOI:10.1371/journal.pone.0158447 |
| [19] | Yang S, Du GC, Chen J, et al. Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis. Appl Microbiol Biotechnol, 2017, 101(10): 4151-4161. DOI:10.1007/s00253-017-8142-7 |
| [20] | Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics, 1999, 15(7–8): 607-611. |
| [21] | Liu Q, Wu KY, Cheng YB, et al. Engineering an iterative polyketide pathway in Escherichia coli results in single-form alkene and alkane overproduction. Metab Eng, 2005, 28: 82-90. |
| [22] | K?nig L, Hartz P, Bernhardt R, et al. High-yield C11-oxidation of hydrocortisone by establishment of an efficient whole-cell system in Bacillus megaterium. Metab Eng, 2019, 55: 59-67. DOI:10.1016/j.ymben.2019.06.005 |
| [23] | De Mey M, Maertens J, Lequeux GJ, et al. . Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol, 2007, 7: 34. DOI:10.1186/1472-6750-7-34 |
| [24] | Braatsch S, Helmark S, Kranz H, et al. Escherichia coli strains with promoter libraries constructed by Red/ET recombination pave the way for transcriptional fine-tuning. Biotechniques, 2008, 45(3): 335-337. DOI:10.2144/000112907 |
| [25] | Wei L, Xu N, Wang YR, et al. Promoter library-based module combination (PLMC) technology for optimization of threonine biosynthesis in Corynebacterium glutamicum. Appl Microbiol Biotechnol, 2018, 102(9): 4117-4130. DOI:10.1007/s00253-018-8911-y |
| [26] | Nevoigt E, Kohnke J, Fischer CR, et al. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol, 2006, 72(8): 5266-5273. DOI:10.1128/AEM.00530-06 |
| [27] | Qin XL, Qian JC, Yao GF, et al. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl Environ Microbiol, 2011, 77(11): 3600-3608. DOI:10.1128/AEM.02843-10 |
| [28] | Hartner FS, Ruth C, Langenegger D, et al. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res, 2008, 36(12): e76. DOI:10.1093/nar/gkn369 |
| [29] | Liu DY, Mao ZT, Guo JX, et al. Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synth Biol, 2018, 7(7): 1785-1797. DOI:10.1021/acssynbio.8b00115 |
| [30] | Li TT, Li T, Ji WY, et al. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp. Biotechnol J, 2016, 11(2): 219-227. DOI:10.1002/biot.201400828 |
| [31] | Jiao S, Li X, Yu HM, et al. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol Bioeng, 2017, 114(4): 832-842. DOI:10.1002/bit.26197 |
| [32] | Wu SM, Feng C, Zhong J, et al. Enhanced production of recombinant nattokinase in Bacillus subtilis by promoter optimization. World J Microbiol Biotchnol, 2011, 27(1): 99-106. DOI:10.1007/s11274-010-0432-5 |
| [33] | Blazeck J, Liu LQ, Redden H, et al. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol, 2011, 77(22): 7905-7914. DOI:10.1128/AEM.05763-11 |
| [34] | Blazeck J, Garg R, Reed B, et al. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol Bioeng, 2012, 109(11): 2884-2895. DOI:10.1002/bit.24552 |
| [35] | Blazeck J, Miller J, Pan A, et al. Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Appl Microbiol Biotechnol, 2014, 98(19): 8155-8164. DOI:10.1007/s00253-014-5895-0 |
| [36] | Madzak C, Tréton B, Blanchin-Roland S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J Mol Microbiol Biotechnol, 2000, 2(2): 207-216. |
| [37] | ?ztürk S, Ergün BG, ?al?k P. Double promoter expression systems for recombinant protein production by industrial microorganisms. Appl Microbiol Biotechnol, 2017, 101(20): 7459-7475. DOI:10.1007/s00253-017-8487-y |
| [38] | Zhang K, Su LQ, Duan XG, et al. High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb Cell Fact, 2017, 16(1): 32. DOI:10.1186/s12934-017-0649-1 |
| [39] | Widner B, Thomas M, Sternberg D, et al. Development of marker-free strains of Bacillus subtilis capable of secreting high levels of industrial enzymes. J Ind Microbiol Biotechnol, 2000, 25(4): 204-212. DOI:10.1038/sj.jim.7000051 |
| [40] | Kang HK, Jang JH, Shim JH, et al. Efficient constitutive expression of thermostable 4-α-glucanotransferase in Bacillus subtilis using dual promoters. World J Microb Biotechnol, 2010, 26(10): 1915-1918. DOI:10.1007/s11274-010-0351-5 |
| [41] | 刘刚, 张燕, 邢苗. 双启动子对重组溶源性枯草杆菌中外源蛋白表达的增强作用. 生物工程学报, 2006, 22(2): 191-197. Liu G, Zhang Y, Xing M. Dual promoters enhance heterologous enzyme production from bacterial phage based recombinant Bacillus subtilis. Chin J Biotech, 2006, 22(2): 191-197 (in Chinese). DOI:10.3321/j.issn:1000-3061.2006.02.004 |
| [42] | Liu SL, Du K. Enhanced expression of an endoglucanase in Bacillus subtilis by using the sucrose-inducible sacB promoter and improved properties of the recombinant enzyme. Protein Expr Purif, 2012, 83(2): 164-168. DOI:10.1016/j.pep.2012.03.015 |
| [43] | Guan CR, Cui WJ, Cheng JT, et al. Construction of a highly active secretory expression system via an engineered dual promoter and a highly efficient signal peptide in Bacillus subtilis. New Biotechnol, 2016, 33(3): 372-379. DOI:10.1016/j.nbt.2016.01.005 |
| [44] | Kinney JB, Murugan A, Callan CG Jr, et al. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA, 2010, 107(20): 9158-9163. DOI:10.1073/pnas.1004290107 |
| [45] | Brewster RC, Jones DL, Phillips R. Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput Biol, 2012, 8(12): e1002811. DOI:10.1371/journal.pcbi.1002811 |
| [46] | Xu YQ, Chu HP, Gao C, et al. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2, 3-butanediol. Metab Eng, 2014, 23: 22-33. DOI:10.1016/j.ymben.2014.02.004 |
| [47] | Sun J, Shao ZY, Zhao H, et al. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol Bioeng, 2012, 109(8): 2082-2092. DOI:10.1002/bit.24481 |
| [48] | Yuan LZ, Rouvière PE, LaRossa RA, et al. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng, 2006, 8(1): 79-90. DOI:10.1016/j.ymben.2005.08.005 |
| [49] | Luo ZW, Kim WJ, Lee SY. Metabolic engineering of Escherichia coli for efficient production of 2-pyrone-4, 6-dicarboxylic acid from glucose. ACS Synth Biol, 2018, 7(9): 2296-2307. DOI:10.1021/acssynbio.8b00281 |
| [50] | Jung YK, Kim TY, Park SJ, et al. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol Bioeng, 2010, 105(1): 161-171. DOI:10.1002/bit.22548 |
| [51] | Atsumi S, Cann AF, Connor MR, et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng, 2008, 10(6): 305-311. DOI:10.1016/j.ymben.2007.08.003 |
| [52] | Ling MX, Liu YF, Li JH, et al. Combinatorial promoter engineering of glucokinase and phosphoglucoisomerase for improved N-acetylglucosamine production in Bacillus subtilis. Bioresour Technol, 2017, 245: 1093-1102. DOI:10.1016/j.biortech.2017.09.063 |
| [53] | Becker J, Zelder O, H?fner S, et al. From zero to hero-design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab Eng, 2011, 13(2): 159-168. DOI:10.1016/j.ymben.2011.01.003 |
| [54] | Shukal S, Chen XX, Zhang CQ. Systematic engineering for high-yield production of viridiflorol and amorphadiene in auxotrophic Escherichia coli. Metab Eng, 2019, 55: 170-178. DOI:10.1016/j.ymben.2019.07.007 |
| [55] | Zhou L, Ding Q, Jiang GZ, et al. Chromosome engineering of Escherichia coli for constitutive production of salvianic acid A. Microb Cell Fact, 2017, 16(1): 84. DOI:10.1186/s12934-017-0700-2 |
| [56] | Wang HJ, Liu WQ, Shi F, et al. Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli. Metab Eng, 2018, 48: 279-287. DOI:10.1016/j.ymben.2018.06.007 |
| [57] | Wu JJ, Du GC, Zhou JW, et al. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng, 2013, 16: 48-55. DOI:10.1016/j.ymben.2012.11.009 |
| [58] | Wu JJ, Liu PR, Fan YM, et al. Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from L-tyrosine. J Biotechnol, 2013, 167(4): 404-411. DOI:10.1016/j.jbiotec.2013.07.030 |
| [59] | Wu JJ, Zhou TT, Du GC, et al. Modular optimization of heterologous pathways for de novo synthesis of (2S)-naringenin in Escherichia coli. PLoS ONE, 2014, 9(7): e101492. DOI:10.1371/journal.pone.0101492 |
| [60] | Venayak N, Anesiadis N, Cluett WR, et al. Engineering metabolism through dynamic control. Curr Opin Biotechnol, 2015, 34: 142-152. DOI:10.1016/j.copbio.2014.12.022 |
| [61] | Nakashima N, Akita H, Hoshino T. Establishment of a novel gene expression method, BICES (biomass-inducible chromosome-based expression system), and its application to the production of 2, 3-butanediol and acetoin. Metab Eng, 2014, 25: 204-214. DOI:10.1016/j.ymben.2014.07.011 |
| [62] | Kim S, Lee K, Bae SJ, et al. Promoters inducible by aromatic amino acids and γ-aminobutyrate (GABA) for metabolic engineering applications in Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 2015, 99(6): 2705-2714. DOI:10.1007/s00253-014-6303-5 |
| [63] | Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene, 1994, 151(1/2): 131-135. |
| [64] | Zhou L, Niu DD, Tian KM, et al. Genetically switched D-lactate production in Escherichia coli. Metab Eng, 2012, 14(5): 560-568. DOI:10.1016/j.ymben.2012.05.004 |
| [65] | Rajkumar AS, Liu GD, Bergenholm D, et al. Engineering of synthetic, stress-responsive yeast promoters. Nucleic Acids Res, 2016, 44(17): e136. DOI:10.1093/nar/gkw553 |
| [66] | Yin X, Shin HD, Li JH, et al. P gas, A low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger. Appl Environ Microbiol, 2017, 83(6): e03222-16. |
| [67] | Senoo S, Tandar ST, Kitamura S, et al. Light-inducible flux control of triosephosphate isomerase on glycolysis in Escherichia coli. Biotechnol Bioeng, 2019, 116(12): 3292-3300. DOI:10.1002/bit.27148 |
| [68] | Tan SZ, Prather KLJ. Dynamic pathway regulation: recent advances and methods of construction. Curr Opin Chem Biol, 2017, 41: 28-35. DOI:10.1016/j.cbpa.2017.10.004 |
| [69] | Yuan JF, Ching CB. Dynamic control of ERG9 expression for improved amorpha-4, 11-diene production in Saccharomyces cerevisiae. Microb Cell Fact, 2015, 14: 38. DOI:10.1186/s12934-015-0220-x |
| [70] | Raman S, Rogers JK, Taylor ND, et al. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA, 2014, 111(50): 17803-17808. DOI:10.1073/pnas.1409523111 |
| [71] | Younger AKD, Dalvie NC, Rottinghaus AG, et al. Engineering modular biosensors to confer metabolite-responsive regulation of transcription. ACS Synth Biol, 2017, 6(2): 311-325. DOI:10.1021/acssynbio.6b00184 |
| [72] | Xu XH, Li XL, Liu YF, et al. Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat Chem Biol, 2020, 16(11): 1261-1268. DOI:10.1038/s41589-020-0637-3 |
| [73] | Xu P, Li LY, Zhang FM, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA, 2014, 111(31): 11299-11304. DOI:10.1073/pnas.1406401111 |
| [74] | Liu D, Xiao Y, Evans BS, et al. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol, 2015, 4(2): 132-140. DOI:10.1021/sb400158w |
| [75] | David F, Nielsen J, Siewers V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae. ACS Synth Biol, 2016, 5(3): 224-233. DOI:10.1021/acssynbio.5b00161 |
| [76] | Jha RK, Kern TL, Fox DT, et al. Engineering an Acinetobacter regulon for biosensing and high-throughput enzyme screening in E. coli via flow cytometry. Nucleic Acids Res, 2014, 42(12): 8150-8160. DOI:10.1093/nar/gku444 |
| [77] | Lo TM, Chng SH, Teo WS, et al. A two-layer gene circuit for decoupling cell growth from metabolite production. Cell Syst, 2016, 3(2): 133-143. DOI:10.1016/j.cels.2016.07.012 |
| [78] | Chen W, Zhang S, Jiang PX, et al. Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metab Eng, 2015, 30: 149-155. DOI:10.1016/j.ymben.2015.05.004 |
| [79] | Liang CN, Zhang XX, Wu JY, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab Eng, 2020, 57: 239-246. DOI:10.1016/j.ymben.2019.12.002 |
| [80] | Adams BL, Carter KK, Guo M, et al. Evolved quorum sensing regulator, LsrR, for altered switching functions. ACS Synth Biol, 2014, 3(4): 210-219. DOI:10.1021/sb400068z |
| [81] | Lindemann A, Pessi G, Schaefer AL, et al. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci USA, 2011, 108(40): 16765-16770. DOI:10.1073/pnas.1114125108 |
| [82] | Hirakawa H, Oda Y, Phattarasukol S, et al. Activity of the Rhodopseudomonas palustris p-coumaroyl-homoserine lactone-responsive transcription factor RpaR. J Bacteriol, 2011, 193(10): 2598-2607. DOI:10.1128/JB.01479-10 |
| [83] | Xue HR, Shi HL, Yu Z, et al. Design, construction, and characterization of a set of biosensors for aromatic compounds. ACS Synth Biol, 2014, 3(12): 1011-1014. DOI:10.1021/sb500023f |
| [84] | Stanton BC, Nielsen AA, Tamsir A, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol, 2014, 10(2): 99-105. DOI:10.1038/nchembio.1411 |
| [85] | O'Rourke S, Wietzorrek A, Fowler K, et al. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol Microbiol, 2009, 71(3): 763-778. DOI:10.1111/j.1365-2958.2008.06560.x |
| [86] | Siedler S, Stahlhut SG, Malla S, et al. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng, 2014, 21: 2-8. DOI:10.1016/j.ymben.2013.10.011 |
| [87] | Chuang JS, Rivoire O, Leibler S. Simpson's paradox in a synthetic microbial system. Science, 2009, 323(5911): 272-275. DOI:10.1126/science.1166739 |
| [88] | Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng, 2008, 10(5): 255-266. DOI:10.1016/j.ymben.2008.06.004 |
| [89] | Wei YP, Ryan GT, Flores-Mireles AL, et al. Saturation mutagenesis of a CepR binding site as a means to identify new quorum-regulated promoters in Burkholderia cenocepacia. Mol Microbiol, 2011, 79(3): 616-632. DOI:10.1111/j.1365-2958.2010.07469.x |
| [90] | Kim EM, Woo HM, Tian T, et al. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng, 2017, 44: 325-336. DOI:10.1016/j.ymben.2017.11.004 |
| [91] | Liu HW, Lu T. Autonomous production of 1, 4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab Eng, 2015, 29: 135-141. DOI:10.1016/j.ymben.2015.03.009 |
| [92] | Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng, 2015, 30: 7-15. DOI:10.1016/j.ymben.2015.04.005 |
| [93] | Williams TC, Nielsen LK, Vickers CE. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol, 2013, 2(3): 136-149. DOI:10.1021/sb300110b |
| [94] | Williams TC, Averesch NJH, Winter G, et al. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng, 2015, 29: 124-134. DOI:10.1016/j.ymben.2015.03.008 |
| [95] | McKeague M, Wong RS, Smolke CD. Opportunities in the design and application of RNA for gene expression control. Nucleic Acids Res, 2016, 44(7): 2987-2999. DOI:10.1093/nar/gkw151 |
| [96] | Leistra AN, Curtis NC, Contreras LM. Regulatory non-coding sRNAs in bacterial metabolic pathway engineering. Metab Eng, 2019, 52: 190-214. DOI:10.1016/j.ymben.2018.11.013 |
| [97] | Zhao Y, Wang CS, Li FF, et al. Targeted optimization of central carbon metabolism for engineering succinate production in Escherichia coli. BMC Biotechnol, 2016, 16: 52. DOI:10.1186/s12896-016-0284-7 |
| [98] | Yang D, Yoo S, Gu CD, et al. Expanded synthetic small regulatory RNA expression platforms for rapid and multiplex gene expression knockdown. Metab Eng, 2019, 54: 180-190. DOI:10.1016/j.ymben.2019.04.003 |
| [99] | Yang D, Kim WJ, Yoo SM, et al. Repurposing type Ⅲ polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci USA, 2018, 115(4): 9835-9844. |
| [100] | Yoo SM, Na D, Lee SY. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat Protoc, 2013, 8(9): 1694-1707. DOI:10.1038/nprot.2013.105 |
| [101] | Noh M, Yoo SM, Kim WJ, et al. Gene expression knockdown by modulating synthetic small RNA expression in Escherichia coli. Cell Syst, 2017, 5(4): 418-426. DOI:10.1016/j.cels.2017.08.016 |
| [102] | Sung M, Yoo SM, Jun R, et al. Optimization of phage λ promoter strength for synthetic small regulatory RNA-based metabolic engineering. Biotechnol Bioproc Eng, 2016, 21(4): 483-490. DOI:10.1007/s12257-016-0245-y |
| [103] | Sun DH, Chen JZ, Wang Y, et al. Metabolic engineering of Corynebacterium glutamicum by synthetic small regulatory RNAs. J Ind Microbiol Biotechnol, 2019, 46(2): 203-208. DOI:10.1007/s10295-018-02128-4 |
| [104] | Cao JC, Arha M, Sudrik C, et al. A universal strategy for regulating mRNA translation in prokaryotic and eukaryotic cells. Nucleic Acids Res, 2015, 43(8): 4353-4362. DOI:10.1093/nar/gkv290 |
| [105] | Muranaka N, Sharma V, Nomura Y, et al. An efficient platform for genetic selection and screening of gene switches in Escherichia coli. Nucleic Acids Res, 2009, 37(5): e39. DOI:10.1093/nar/gkp039 |
| [106] | Günzelh C, Kühnl F, Arnold K, et al. Beyond plug and pray: context sensitivity and in silico design of artificial neomycin riboswitches. RNA Biol, 2020. DOI:10.1080/15476286.2020.1816336 |
| [107] | Boumezbeur AH, Bruer M, Stoecklin G, et al. Rational engineering of transcriptional riboswitches leads to enhanced metabolite levels in Bacillus subtilis. Metab Eng, 2020, 61: 58-68. DOI:10.1016/j.ymben.2020.05.002 |
| [108] | Zhou LB, Zeng AP. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum. ACS Synth Biol, 2015, 4(6): 729-734. DOI:10.1021/sb500332c |
| [109] | Zhou LB, Zeng AP. Engineering a lysine-ON riboswitch for metabolic control of lysine production in Corynebacterium glutamicum. ACS Synth Biol, 2015, 4(12): 1335-1340. DOI:10.1021/acssynbio.5b00075 |
| [110] | Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet, 2010, 11(3): 181-190. DOI:10.1038/nrg2749 |
| [111] | Lv L, Ren YL, Chen JC, et al. Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: controllable P(3HB-co-4HB) biosynthesis. Metab Eng, 2015, 29: 160-168. DOI:10.1016/j.ymben.2015.03.013 |
| [112] | Li QY, Zhao P, Yin H, et al. CRISPR interference-guided modulation of glucose pathways to boost aconitic acid production in Escherichia coli. Microb Cell Fact, 2020, 19(1): 174. DOI:10.1186/s12934-020-01435-9 |
| [113] | Kim SK, Han GH, Seong W, et al. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab Eng, 2016, 38: 228-240. DOI:10.1016/j.ymben.2016.08.006 |
| [114] | Cleto S, Jensen J, Wendisch VF, et al. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol, 2016, 5(5): 375-385. DOI:10.1021/acssynbio.5b00216 |
| [115] | Westbrook A, Ren X, Oh J, et al. Metabolic engineering to enhance heterologous production of hyaluronic acid in Bacillus subtilis. Metab Eng, 2018, 47: 401-413. DOI:10.1016/j.ymben.2018.04.016 |
| [116] | Zhang JL, Peng YZ, Liu D, et al. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb Cell Fact, 2018, 17(1): 62. DOI:10.1186/s12934-018-0909-8 |
| [117] | Ji XY, Zhao HW, Zhu H, et al. CRISPRi/dCpf1-mediated dynamic metabolic switch to enhance butenoic acid production in Escherichia coli. Appl Microbiol Biotechnol, 2020, 104(12): 5385-5393. DOI:10.1007/s00253-020-10610-2 |
| [118] | Li D, Lv L, Chen JC, et al. Controlling microbial PHB synthesis via CRISPRi. Appl Microbiol Biotechnol, 2017, 101(14): 5861-5867. DOI:10.1007/s00253-017-8374-6 |
| [119] | Zhang XC, Guo YY, Liu X, et al. Engineering cell wall synthesis mechanism for enhanced PHB accumulation in E. coli. Metab Eng, 2018, 45: 32-42. DOI:10.1016/j.ymben.2017.11.010 |
| [120] | Elhadi D, Lv L, Jiang XR, et al. CRISPRi engineering E. coli for morphology diversification. Metab Eng, 2016, 38: 358-369. DOI:10.1016/j.ymben.2016.09.001 |
| [121] | Chang YZ, Su TY, Qi QS, et al. Easy regulation of metabolic flux in Escherichia coli using an endogenous type Ⅰ-E CRISPR-Cas system. Microb Cell Fact, 2016, 15: 195. DOI:10.1186/s12934-016-0594-4 |
| [122] | Wu MY, Sung LY, Li H, et al. Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1, 4-BDO biosynthesis. ACS Synth Biol, 2017, 6(12): 2353-2361. |
| [123] | Kim SK, Seong W, Han GH, et al. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Fact, 2017, 16(1): 188. DOI:10.1186/s12934-017-0802-x |
| [124] | Su TY, Guo Q, Zheng Y, et al. Fine-tuning of hemB using CRISPRi for increasing 5-aminolevulinic acid production in Escherichia coli. Front. Microbiol, 2019, 10: 1731. DOI:10.3389/fmicb.2019.01731 |
| [125] | Gao C, Wang SH, Hu GP, et al. Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning. Biotechnol Bioeng, 2018, 115(3): 661-672. DOI:10.1002/bit.26486 |
| [126] | Tao S, Qian Y, Wang X, et al. Regulation of ATP levels in Escherichia coli using CRISPR interference for enhanced pinocembrin production. Microb Cell Fact, 2018, 17: 147. DOI:10.1186/s12934-018-0995-7 |
| [127] | Cai FJ, Guo LQ, Lin JF, et al. A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J Microbiol Biotechnol, 2016, 32(6): 102. DOI:10.1007/s11274-016-2062-z |
| [128] | Sander T, Wang CY, Glatter T, et al. CRISPRi-based downregulation of transcriptional feedback improves growth and metabolism of arginine overproducing E. coli. ACS Synth Biol, 2019, 8(9): 1983-1990. DOI:10.1021/acssynbio.9b00183 |
| [129] | Wang CY, Cao YX, Wang YP, et al. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb Cell Fact, 2019, 18: 90. DOI:10.1186/s12934-019-1139-4 |
| [130] | Lu ZH, Yang SH, Yuan X, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis. Nucleic Acids Res, 2019, 47(7): e40. DOI:10.1093/nar/gkz072 |
| [131] | Wu YK, Chen TC, Liu YF, et al. CRISPRi allows optimal temporal control of N-acetylglucosamine bioproduction by a dynamic coordination of glucose and xylose metabolism in Bacillus subtilis. Metab Eng, 2018, 49: 232-241. DOI:10.1016/j.ymben.2018.08.012 |
| [132] | Zhan YY, Xu Y, Zheng PL, et al. Establishment and application of multiplexed CRISPR interference system in Bacillus licheniformis. Appl Microbiol Biotechnol, 2020, 104(1): 391-403. DOI:10.1007/s00253-019-10230-5 |
| [133] | Park J, Shin H, Lee SM, et al. RNA-guided single/double gene repressions in Corynebacterium glutamicum using an efficient CRISPR interference and its application to industrial strain. Microb Cell Fact, 2018, 17(1): 4. DOI:10.1186/s12934-017-0843-1 |
| [134] | Zhang B, Liu ZQ, Liu C, et al. Application of CRISPRi in Corynebacterium glutamicum for shikimic acid production. Biotechnol Lett, 2016, 38(12): 2153-2161. DOI:10.1007/s10529-016-2207-z |
| [135] | Gauttam R, Seibold GM, Mueller P, et al. A simple dual-inducible CRISPR interference system for multiple gene targeting in Corynebacterium glutamicum. Plasmid, 2019, 103: 25-35. DOI:10.1016/j.plasmid.2019.04.001 |
| [136] | Li Z, Shen YP, Jiang XL, et al. Metabolic evolution and a comparative omics analysis of Corynebacterium glutamicum for putrescine production. J Ind Microbiol Biotechnol, 2018, 45(2): 123-139. DOI:10.1007/s10295-018-2003-y |
| [137] | Tong YJ, Charusanti P, Zhang LX, et al. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol, 2015, 4(9): 1020-1029. DOI:10.1021/acssynbio.5b00038 |
| [138] | Liu X, Gallay C, Kjos M, et al. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol, 2017, 13(5): 931. DOI:10.15252/msb.20167449 |
| [139] | Tan SZ, Reisch CR, Prather KLJ. A robust CRISPR interference gene repression system in Pseudomonas. J Bacteriol, 2018, 200(7): e00575-17. |
| [140] | Delihas N. Regulation of gene expression by trans-encoded antisense RNAs. Mol Microbiol, 1995, 15(3): 411-414. DOI:10.1111/j.1365-2958.1995.tb02254.x |
| [141] | Saberi F, Kamali M, Najafi A, et al. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett, 2016, 21: 6. DOI:10.1186/s11658-016-0007-z |
| [142] | Hoynes-O'Connor A, Moon TS. Development of design rules for reliable antisense RNA behavior in E. coli. ACS Synth Biol, 2016, 5(12): 1441-1454. DOI:10.1021/acssynbio.6b00036 |
| [143] | Lioliou E, Romilly C, Romby P, et al. RNA-mediated regulation in bacteria: from natural to artificial systems. New Biotechnol, 2010, 27(3): 222-235. DOI:10.1016/j.nbt.2010.03.002 |
| [144] | Kim JYH, Cha HJ. Down-regulation of acetate pathway through antisense strategy in Escherichia coli: improved foreign protein production. Biotechnol Bioeng, 2003, 83(7): 841-853. DOI:10.1002/bit.10735 |
| [145] | Solomon KV, Sanders TM, Prather KLJ. A dynamic metabolite valve for the control of central carbon metabolism. Metab Eng, 2012, 14(6): 661-671. DOI:10.1016/j.ymben.2012.08.006 |
| [146] | Kang Z, Wang XR, Li KY, et al. Small RNA RyhB as a potential tool used for metabolic engineering in Escherichia coli. Biotechnol Lett, 2012, 34(3): 527-531. DOI:10.1007/s10529-011-0794-2 |
| [147] | Meng HL, Xiong ZQ, Song SJ, et al. Construction of polyketide overproducing Escherichia coli strains via synthetic antisense RNAs based on in silico fluxome analysis and comparative transcriptome analysis. Biotechnol J, 2016, 11(4): 530-541. DOI:10.1002/biot.201500351 |
| [148] | Yang YP, Lin YH, Li LY, et al. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng, 2015, 29: 217-226. DOI:10.1016/j.ymben.2015.03.018 |
| [149] | Wang MM, Yu HM, Shen ZY. Antisense RNA-based strategy for enhancing surfactin production in Bacillus subtilis TS1726 via overexpression of the unconventional biotin carboxylase Ⅱ to enhance ACCase activity. ACS Synth Biol, 2019, 8(2): 251-256. DOI:10.1021/acssynbio.8b00459 |
| [150] | Biedendieck R, Malten M, Barg H, et al. Metabolic engineering of cobalamin (vitamin B12) production in Bacillus megaterium. Microb Biotechnol, 2010, 3(1): 24-37. DOI:10.1111/j.1751-7915.2009.00125.x |
| [151] | Desai RP, Papoutsakis ET. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl Environ Microbiol, 1999, 65(3): 936. DOI:10.1128/AEM.65.3.936-945.1999 |
| [152] | Tummala SB, Welker NE, Papoutsakis ET. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol, 2003, 185(6): 1923-1934. DOI:10.1128/JB.185.6.1923-1934.2003 |
| [153] | Steitz JA. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature, 1969, 224(5223): 957-964. DOI:10.1038/224957a0 |
| [154] | Chen HY, Bjerknes M, Kumar R, et al. Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli m RNAs. Nucleic Acids Res, 1994, 22(23): 4953-4957. DOI:10.1093/nar/22.23.4953 |
| [155] | De Smit MH, Duin JV. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA, 1990, 87(19): 7668-7672. DOI:10.1073/pnas.87.19.7668 |
| [156] | Seo SW, Yang JS, Cho HS, et al. Predictive combinatorial design of mRNA translation initiation regions for systematic optimization of gene expression levels. Sci Rep, 2014, 4: 4515. |
| [157] | Wang JP, Ai XL, Mei H, et al. High-throughput identification of promoters and screening of highly active promoter-5?-UTR DNA region with different characteristics from Bacillus thuringiensis. PLoS ONE, 2013, 8(5): e62960. DOI:10.1371/journal.pone.0062960 |
| [158] | Na D, Lee D. RBSDesigner: software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics, 2010, 26(20): 2633-2634. DOI:10.1093/bioinformatics/btq458 |
| [159] | Grünberg R, Serrano L. Strategies for protein synthetic biology. Nucleic Acids Res, 2010, 38(8): 2663-2675. DOI:10.1093/nar/gkq139 |
| [160] | Li X, Xu SQ, Zhang X, et al. Design of a high-efficiency synthetic system for L-asparaginase production in Bacillus subtilis. Eng Life Sci, 2019, 19(3): 229-239. DOI:10.1002/elsc.201800166 |
| [161] | Bujara M, Panke S. Engineering in complex systems. Curr Opin Biotechnol, 2010, 21(5): 586-591. DOI:10.1016/j.copbio.2010.07.007 |
| [162] | Xu P, Gu Q, Wang WY, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun, 2013, 4: 1409. DOI:10.1038/ncomms2425 |
| [163] | Wang B, Hu QT, Zhang Y, et al. A RecET-assisted CRISPR-Cas9 genome editing in Corynebacterium glutamicum. Microb Cell Fact, 2018, 17(1): 63. DOI:10.1186/s12934-018-0910-2 |
| [164] | Liu Y, Zhu LF, Qi WP, et al. Biocatalytic production of D-p-hydroxyphenylglycine by optimizing protein expression and cell wall engineering in Escherichia coli. Appl Microbiol Biotechnol, 2019, 103(21/22): 8839-8851. DOI:10.1007/s00253-019-10155-z |
| [165] | Looman AC, Bodlaender JM, Comstock LJ, et al. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J, 1987, 6(8): 2489-2492. DOI:10.1002/j.1460-2075.1987.tb02530.x |
| [166] | Aiba H, Mori K, Tanaka M, et al. The complete nucleotide sequence of the adenylate cyclase gene of Escherichia coli. Nucleic Acids Res, 1984, 12(24): 9427-9440. DOI:10.1093/nar/12.24.9427 |
| [167] | Danchin A, Guiso N, Roy A, et al. Identification of the Escherichia coli cya gene product as authentic adenylate cyclase. J Mol Biol, 1984, 175(3): 403-408. DOI:10.1016/0022-2836(84)90356-5 |
| [168] | Reddy P, Peterkofsky A, McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci USA, 1985, 82(17): 5656-5660. DOI:10.1073/pnas.82.17.5656 |
| [169] | Wu ZQ, Zhao DD, Li SW, et al. Combinatorial modulation of initial codons for improved zeaxanthin synthetic pathway efficiency in Escherichia coli. MicrobiologyOpen, 2019, 8(12): e930. |
| [170] | Chu D, Kazana E, Bellanger N, et al. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J, 2014, 33(1): 21-34. DOI:10.1002/embj.201385651 |
| [171] | Bonekamp F, Jensen KF. The AGG codon is translated slowly in E. coli even at very low expression levels. Nucleic Acids Res, 1988, 16(7): 3013-3024. DOI:10.1093/nar/16.7.3013 |
| [172] | Rosenberg AH, Goldman E, Dunn JJ, et al. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol, 1993, 175(3): 716-722. DOI:10.1128/JB.175.3.716-722.1993 |
| [173] | Chen GT, Inouye M. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev, 1994, 8(21): 2641-2652. DOI:10.1101/gad.8.21.2641 |
| [174] | Stenstr?m CM, Jin HN, Major LL, et al. Codon bias at the 3'-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene, 2001, 263(1/2): 273-284. |
| [175] | Finger C, Gamer M, Klunkelfu? S, et al. Impact of rare codons and the functional coproduction of rate-limiting tRNAs on recombinant protein production in Bacillus megaterium. Appl Microbiol Biotechnol, 2015, 99(21): 8999-9010. DOI:10.1007/s00253-015-6744-5 |
| [176] | 冯凡, 许杨, 陶勇, 等. 提高大肠杆菌通过MVA途径合成异戊二烯. 生物工程学报, 2015, 31(7): 1073-1081. Feng F, Xu Y, Tao Y, et al. Improving isoprene production by engineered heterologous mevalonate pathway in Escherichia coli. Chin J Biotech, 2015, 31(7): 1073-1081 (in Chinese). |
| [177] | Berg OG, Kurland CG. Growth rate-optimised tRNA abundance and codon usage. J Mol Biol, 1997, 270(4): 544-550. DOI:10.1006/jmbi.1997.1142 |
| [178] | Makoff AJ, Oxer MD, Romanos MA, et al. Expression of tetanus toxin fragment C in E. coli: high level expression by removing rare codons. Nucleic Acids Res, 1989, 17(24): 10191-10202. DOI:10.1093/nar/17.24.10191 |
| [179] | Hu XY, Shi QW, Yang T, et al. Specific replacement of consecutive AGG codons results in high-level expression of human cardiac troponin T in Escherichia coli. Protein Expr Purif, 1996, 7(3): 289-293. DOI:10.1006/prep.1996.0041 |
| [180] | Johansson AS, Bolton-Grob R, Mannervik B. Use of silent mutations in cDNA encoding human glutathione transferase M2-2 for optimized expression in Escherichia coli. Protein Expr Purif, 1999, 17(1): 105-112. DOI:10.1006/prep.1999.1117 |
| [181] | Li AL, Kato Z, Ohnishi H, et al. Optimized gene synthesis and high expression of human interleukin-18. Protein Expr Purif, 2003, 32(1): 110-118. DOI:10.1016/j.pep.2003.08.003 |
| [182] | Kotula L, Curtis PJ. Evaluation of foreign gene codon optimization in yeast: expression of a mouse IG kappa chain. Bio/Technology, 1991, 9(12): 1386-1389. DOI:10.1038/nbt1291-1386 |
| [183] | Batard Y, Hehn A, Nedelkina S, et al. Increasing expression of P450 and P450-reductase proteins from monocots in heterologous systems. Arch Biochem Biophys, 2000, 379(1): 161-169. DOI:10.1006/abbi.2000.1867 |
| [184] | Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet, 2014, 15(3): 205-213. DOI:10.1038/nrg3645 |
| [185] | Pop C, Rouskin S, Ingolia NT, et al. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol, 2014, 10(12): 770. DOI:10.15252/msb.20145524 |
| [186] | Qian WF, Yang JR, Pearson NM, et al. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet, 2012, 8(3): e1002603. DOI:10.1371/journal.pgen.1002603 |
| [187] | Dana A, Tuller T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res, 2014, 42(14): 9171-9181. DOI:10.1093/nar/gku646 |
| [188] | Gardin J, Yeasmin R, Yurovsky A, et al. Measurement of average decoding rates of the 61 sense codons in vivo. Elife, 2014, 3: e03735. DOI:10.7554/eLife.03735 |
| [189] | Fritze J, Zhang MY, Luo Q, et al. An overview of the bacterial SsrA system modulating intracellular protein levels and activities. Appl Microbiol Biotechnol, 2020, 104(12): 5229-5241. DOI:10.1007/s00253-020-10623-x |
| [190] | Gao C, Hou JS, Xu P, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat Commun, 2019, 10(1): 3751. DOI:10.1038/s41467-019-11793-7 |
| [191] | Tandar ST, Senoo S, Toya Y, et al. Optogenetic switch for controlling the central metabolic flux of Escherichia coli. Metab Eng, 2019, 55: 68-75. DOI:10.1016/j.ymben.2019.06.002 |
| [192] | Moser F, Espah BA, Ghodasara AN, et al. Dynamic control of endogenous metabolism with combinatorial logic circuits. Mol Syst Biol, 2018, 14(11): e8605. |
| [193] | Torella JP, Ford TJ, Kim SN, et al. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc Natl Acad Sci USA, 2013, 110(28): 11290-11295. DOI:10.1073/pnas.1307129110 |
| [194] | Hou JS, Gao C, Guo L, et al. Rewiring carbon flux in Escherichia coli using a bifunctional molecular switch. Metab Eng, 2020, 61: 47-57. DOI:10.1016/j.ymben.2020.05.004 |
| [195] | Keiler KC. Physiology of tmRNA: what gets tagged and why?. Curr Opin Microbiol, 2007, 10(2): 169-175. DOI:10.1016/j.mib.2007.03.014 |
| [196] | Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 1996, 271(5251): 990-993. DOI:10.1126/science.271.5251.990 |
| [197] | McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell, 2006, 22(5): 701-707. DOI:10.1016/j.molcel.2006.04.027 |
| [198] | Brockman IM, Prather KLJ. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites. Metab Eng, 2015, 28: 104-113. DOI:10.1016/j.ymben.2014.12.005 |
| [199] | Reizman IMB, Stenger AR, Reisch CR, et al. mprovement of glucaric acid production in E. coli via dynamic control of metabolic fluxes. Metab Eng Commun, 2015, 2: 109-116. DOI:10.1016/j.meteno.2015.09.002 |
| [200] | Durante-Rodríguez G, De Lorenzo V, Nikel PI. A post-translational metabolic switch enables complete decoupling of bacterial growth from biopolymer production in engineered Escherichia coli. ACS Synth Biol, 2018, 7(11): 2686-2697. DOI:10.1021/acssynbio.8b00345 |
| [201] | Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol, 2008, 70(4): 1012-1025. |
| [202] | Boyle PM, Silver PA. Parts plus pipes: synthetic biology approaches to metabolic engineering. Metab Eng, 2012, 14(3): 223-232. DOI:10.1016/j.ymben.2011.10.003 |
| [203] | Davidson EA, Windram OPF, Bayer TS. Building synthetic systems to learn nature's design principles//Soyer O, Ed. Evolutionary Systems Biology. New York, NY: Springer, 2012, 751: 411-429. |
| [204] | Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metab Eng, 2012, 14(3): 189-195. DOI:10.1016/j.ymben.2012.01.004 |
| [205] | Singh V. Recent advancements in synthetic biology: current status and challenges. Gene, 2014, 535(1): 1-11. DOI:10.1016/j.gene.2013.11.025 |
| [206] | 刘波, 陶勇. 生物制造"细胞工厂"的设计与组装. 生物工程学报, 2019, 35(10): 1942-1954. Liu B, Tao Y. Design and assembly of bio-manufacturing "cell factory". Chin J Biotech, 2019, 35(10): 1942-1954 (in Chinese). |
| [207] | Egbert RG, Klavins E. Fine-tuning gene networks using simple sequence repeats. Proc Natl Acad Sci USA, 2012, 109(42): 16817-16822. DOI:10.1073/pnas.1205693109 |
| [208] | Lucks JB, Qi L, Mutalik VK, et al. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci USA, 2011, 108(21): 8617-8622. DOI:10.1073/pnas.1015741108 |
| [209] | Herai S, Hashimoto Y, Higashibata H, et al. Hyper-inducible expression system for streptomycetes. Proc Natl Acad Sci USA, 2004, 101(39): 14031-14035. DOI:10.1073/pnas.0406058101 |
| [210] | Zhang J, Liu YJ, Cui GZ, et al. A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum. Biotechnol Biofuels, 2015, 8: 36. DOI:10.1186/s13068-015-0214-2 |
| [211] | Yansura DG, Henner DJ. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA, 1984, 81(2): 439-443. DOI:10.1073/pnas.81.2.439 |
| [212] | Weinmann P, Gossen M, Hillen W, et al. A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J, 1994, 5(4): 559-569. DOI:10.1046/j.1365-313X.1994.5040559.x |
| [213] | Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl, 2008, 47(14): 2604-2607. DOI:10.1002/anie.200703700 |
| [214] | Ceroni F, Algar R, Stan GB, et al. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods, 2015, 12(5): 415-418. DOI:10.1038/nmeth.3339 |
