
 , 王泽昊1, 王婷月1
, 王泽昊1, 王婷月1 1. 沈阳农业大学 植物保护学院,辽宁 沈阳 110866;
2. 阿尔伯塔省农业部 植物健康实验室,加拿大阿尔伯塔 埃德蒙顿 T5Y 6H3
收稿日期:2018-09-08;接收日期:2018-12-04;网络出版时间:2019-01-08
基金项目:沈阳农业大学引进人才科研启动费项目(No. 20153040),辽宁省高等学校优秀人才支持计划(No. LR2015058)资助
摘要:核盘菌Sclerotinia sclerotiorum是一种典型的死体营养型植物病原真菌,全球分布且寄主范围广泛,严重危害多种植物,对农业生产造成严重损失。核盘菌研究主要集中在真菌生物学及病理学等方面。近年来,随着高通量分析技术的不断改进,多种组学技术为系统生物学研究提供了平台。文中主要综述利用多种组学研究方法在植物病原真菌核盘菌研究中的应用及研究进展,探讨开展植物病原物及病害发展的系统性研究思路,以期为核盘菌的分子生物学及致病机理等研究提供参考,同时也为其他植物病原物及病害系统研究提供理论依据。
关键词:核盘菌草酸组学致病机理分子互作
Application of omics approaches in Sclerotinia sclerotiorum
Huiying Sun1, Jie Feng2, Yue Liang1

 , Zehao Wang1, Tingyue Wang1
, Zehao Wang1, Tingyue Wang1 1. College of Plant Protection, Shenyang Agricultural University, Shenyang 110866, Liaoning, China;
2. 2 The Alberta Plant Health Lab, Alberta Agriculture and Forestry, Edmonton, Alberta T5Y 6H3, Canada
Received: September 8, 2018; Accepted: December 4, 2018; Published: January 8, 2019
Supported by: Scientific Research Foundation for the Introduced Talents of Shenyang Agricultural University (No. 20153040), Program for Liaoning Excellent Talents in University (No. LR2015058)
Corresponding author: Yue Liang. Tel/Fax: +86-24-88487148; E-mail: yliang@syau.edu.cn.
Abstract: Sclerotinia sclerotiorum is a typical necrotrophic plant pathogenic fungus that distributes worldwide and causes severe diseases on a broad-range of plant species. Studies on S. sclerotiorum have been mainly focused on biology and pathology. The development of high-throughput technologies enabled multi-omics approaches for systems biology. This review summarizes current researches on S. sclerotiorum and proposes systemic strategies for understanding its biology and pathology, to provide novel insights and references for further investigation on molecular biology and pathogenesis of the pathogenic fungi and the pathosystems.
Keywords: Sclerotinia sclerotiorumoxalic acidomicspathogenesismolecular interactions
核盘菌是一种典型的死体营养型真菌,在世界各地引起多种病害,属于广泛分布且具毁灭性的植物病原菌之一[1]。核盘菌研究多集中于寄主范围、生物学特性、侵染机制及防控方法等方面。近年来随着分子生物学和生物化学研究方法的发展,多层面信息资源的整合以及多组学(Multi-omics)方法的运用,为探索微生物生物学等方面奠定基础[2]。因此,本文主要综述利用多种组学研究方法在植物病原真菌核盘菌研究中的应用及研究进展,探讨针对植物病原物及病害发展的系统性研究思路,以期为核盘菌的分子生物学及致病机理等研究提供参考,同时也为其他植物病原物及病害系统研究提供理论依据。
1 核盘菌及其重要性核盘菌(Sclerotinia sclerotiorum (Lib.) de Bary)属于真菌界(Fungi)、子囊菌门(Ascomycota)、盘菌纲(Discomycetes)、核盘菌属(Sclerotinia)[1]。核盘菌寄主范围广泛,已知寄主植物超过400种,主要包括双子叶植物(如向日葵、大豆、油菜和多种蔬菜),但部分单子叶植物(如洋葱等)也可被侵染[3]。据统计,该真菌引起病害60余种,包括最为常见的菌核病、棉腐病、软腐病、茎腐病等[1]。核盘菌侵染植物的茎蔓、叶片和果实,造成腐烂坏死;在高湿条件下,罹病部位表面密生棉絮状菌丝体并不断扩大,后期在茎秆及种荚内部形成大量菌核[3]。核盘菌病害循环中越冬菌核在适宜环境下萌发形成子囊盘及子囊,释放大量子囊孢子并随风传播至寄主组织上萌发引起初侵染;随着受侵染的组织(如花瓣)散落在叶片和叶鞘上,造成腐烂坏死,其表面密生白色棉絮状菌丝体,在茎秆及种荚内部产生大量菌核以抵抗外界不利环境并越冬,次年再引起新病害发生[1]。目前,核盘菌引起的病害防控方法有限,尚缺乏高抗品种,生物防治见效缓慢且受到环境限制,而化学防治易造成环境污染并产生抗药性等问题[1]。
2 核盘菌的组学各种组学技术能够高通量定量分析各种生物分子,监测不同生物状态下的变异,已广泛用于微生物系统生物学研究中,包括测定mRNA转录水平的转录组学(Transcriptomics)、定量蛋白丰度的蛋白质组学(Proteomics)、鉴定小分子细胞代谢物的代谢组学(Metabolomics)等[2]。多组学整合生物信息学等方法,为深入了解真菌的生活史和进化、真菌与环境关系(如与寄主植物的互作)等研究提供大量数据与信息[4]。随着核盘菌基因组测序的完成以及基于高通量测序、色谱质谱和核磁共振技术等组学技术的飞速发展,核盘菌分子生物学的研究已经逐步跨入后基因组时代。
2.1 基因组学基因组学(Genomics)作为一门交叉学科,包括基因组的结构、功能和进化等诸多方面[5],其研究对微生物学及生命科学产生了重大影响[6]。其中,真菌基因组计划(1KFG)的开启为真菌遗传与进化的研究提供基础[5]。核盘菌基因组研究始于2005年,并完成初步注释。核盘菌(菌株1980)的基因组约为38.3 Mb (图谱大小39.6 Mb,ASM14694v2);GC含量约40%,低于同类真菌,而外显子比内含子GC含量高6%。基因组含有约1.4万个预测基因,同属于盘菌亚门的其他真菌则含有约1.1万个,这种差异源于其含有大量小于100个氨基酸的预测蛋白[7]。此外,核盘菌的线粒体基因组(NC_035155.1)约为128.8 kb,为深入研究基因组结构组成及进化等提供参考[8]。另外,基因组含有7.7%的重复序列,这可能影响基因组结构及功能[7]。核盘菌编码基因参与真菌发育(如有性生殖、子实体发育及孢子产生)和致病机制(如草酸合成、肽酶的分泌及效应蛋白等),也与进化、代谢、信号途径相关[7]。比较基因组分析预测发现大量分泌蛋白参与植物细胞氧化还原反应等[9],核盘菌通过草酸调控植物氧化还原动态平衡(Redox homeostasis)引起侵染[10]。核盘菌具有非典型的双速基因组(Two-speed genome)特点,能够利用转座和重复序列诱导的点突变(Repeat-induced point mutation,RIP)促进分泌蛋白的突变[11]。
2.2 转录组学转录组学(Transcriptomics)聚焦从RNA水平研究基因表达及调控规律,对揭示真菌与植物的互作及致病性等机制具有重要意义[12]。转录组学研究技术主要包括cDNA文库、表达序列标签文库(Expressed sequence tags,EST)[13]、DNA微阵列技术(DNA microarray)[14]和RNA测序技术(RNA-seq)[15]等。随着转录组技术的兴起,近年来在核盘菌发育和致病等研究中得到了普遍应用。核盘菌的基因组中含有330个转录因子(TFs)基因,69%在cDNA文库或寡核苷酸芯片杂交出现,其中12个转录因子仅在核盘菌中特异性表达[7]。通过比较菌丝及罹病组织的cDNA文库发现,细胞壁降解酶和丝裂原活化蛋白激酶(MAPK)信号途径等参与核盘菌的发育与侵染[16]。罹病植物组织及侵染结构的EST分析表明,不同基因(如草酰乙酸水解酶OAH)在致病过程中的转录表达模式各异[17]。利用EST及microarray分析发现,部分核盘菌预测分泌蛋白参与真菌发育及侵染后寄主的转录调控[9]。另外,通过cDNA抑制消减杂交(SSH)或microarray对核盘菌与寄主植物互作机理研究表明,代谢、信号转导及细胞防御等功能因子参与寄主防卫反应[18-19]。同时,在草酸氧化酶(OxO)转基因大豆microarray分析中也发现抗病反应基因(如细胞色素P450)被诱导表达[20]。此外,非编码RNAs (ncRNAs)包括长链非编码RNA (lncRNAs)和小非编码RNA (sncRNAs),在真核生物中广泛存在并参与转录调控、分化与发育、应激反应等[21]。通过高通量RNA-Seq方法发现lncRNAs参与核盘菌侵染所引起的植物应激反应与调控[22],部分sncRNA参与真菌发育调控[23]。比较不同抗性植物转录组变化揭示大量基因参与病原识别、茉莉酸及乙烯信号及病程相关蛋白等寄主反应[24],特别是草酸能够诱导铁蛋白同源物表达[20]。研究也发现寄主油菜利用乙烯调控氧化还原动态平衡获得诱导抗性[25],并通过水化酶等因子协助病原菌定殖及调控防卫反应[26]。
2.3 蛋白质组学蛋白质组学(Proteomics)是指在大规模水平上研究蛋白质的特征,包括蛋白质的表达、结构和功能等[27]。随着基因组计划及反向遗传学技术的发展,蛋白质组逐渐成为真菌生活史、致病机理与分子互作等领域的主要研究手段[28]。目前,双向凝胶电泳(Two-dimensional gel electrophoresis,2-DE)、质谱(Mass spectrometry, MS)、同位素标记相对和绝对定量技术(Isobaric tags for relative and absolute quantitation,iTRAQ)等逐步用于核盘菌蛋白组学研究[27-29]。应用2-DE和ESI-q-TOF MS/MS首次分析核盘菌蛋白质组发现,菌丝体蛋白主要与代谢和能量产生相关,分泌蛋白具有细胞壁降解酶功能且参与致病[30]。菌丝细胞壁含有大量细胞壁合成酶(如纤维素酶和果胶裂解酶等),但菌核细胞壁蛋白在形成中并未发生明显变化[31]。菌核作为逆境存活结构及病害初侵染源在真菌生活史及病害循环中具有重要作用[1]。通过对不同发育阶段菌核蛋白质组变化分析,脂类及次生代谢功能蛋白参与早期发育,而碳水化合物代谢则在发育后期显著增加[32];菌核渗出液作为菌核发育期主要特征,蛋白组分析表明碳水化合物代谢及信号转导等参与分泌液形成[33]。利用生物信息学方法在基因组水平上预测的分泌蛋白在菌核渗出液及分泌物中均有发现[9, 30, 33]。此外,在蛋白质组水平分析核盘菌侵染的寄主反应发现,光合作用、激素信号及抗氧化等蛋白与植物应激反应相关[34],而氧化还原动态平衡、转录调控等参与植物抗性反应[35],并调控寄主植物细胞程序性死亡[36]。草酸作为核盘菌致病因子介导寄主反应[10],在蛋白质组水平发现能够抑制水杨酸信号途径但不影响NADPH氧化酶参与的氧化还原动态平衡[37],同时通过草酸积累调控不同菌龄的菌丝毒性[38]。
2.4 代谢组学代谢组学(Metabolomics)研究生命活动过程中代谢物、小分子化合物及代谢产物的变化,并寻找代谢物与表型的关联性[39]。真菌次生代谢物的产生过程复杂并与形态发育等相关,例如代谢聚酮化合物(PK)、非核糖体肽(NRPS)、萜烯(Terpenes)和吲哚生物碱(Indole alkaloids)等[40]。核盘菌基因组含有参与氨基酸合成及线粒体功能的初级代谢基因(如NADH脱氢酶),同时具有(非特异性)毒素和其他次级代谢产物关键酶基因[7]。草酸作为非特异性毒素参与核盘菌侵染及寄主反应调控[10],通过高效液相色谱(HPLC)分析发现草酸合成与碳水化合物(如葡萄糖、阿拉伯糖及木聚糖等)和有机酸(琥珀酸、苹果酸、草酰乙酸)等次生代谢物相关[41]。另外,核盘菌含有的生物碱及异香豆素(Isocoumarins)等次生代谢物具有细胞毒素活性[42],聚胺(Polyamine)影响分生孢子及菌核的发育[43-44],选择性毒素核盘菌素(Sclerin)能够引起寄主植物坏死与褪绿[45]。代谢组学研究也有助于阐明植物对核盘菌的抗性机制[46],例如,运用气相色谱-质谱(GC-MS)联用技术发现向日葵抗性与糖类、有机酸、脂肪酸等代谢相关[47];菜豆抗性品种也含有氨基酸和植保素等代谢物[48];苯丙氨酸解氨酶(PAL)活性及植保素异黄酮的变化反映大豆抗病机制差异[49];酚类化合物和木质素参与向日葵防卫反应[50];拟南芥通过脂类代谢参与寄主反应[51];硫代葡萄糖苷和亚麻荠素(Camalexin)也与防卫反应相关[52]。
2.5 功能及病理基因组学功能基因组(Functional genomics)主要基于高通量基因组分析策略在系统水平上研究基因功能[53],病理基因组学(Pathogenomics)则关注病原菌的致病机理及其相关因子的功能与分子互作等[54]。核盘菌通过菌核萌发和有性生殖形成的子囊孢子造成病害发生与流行[55],表明菌核在真菌生活史及病害循环中发挥重要作用[1]。作为典型的同宗配合子囊真菌,交配型基因编码的产物是有性生殖的重要调控因子[55]。核盘菌交配型基因座融合并相连,包括MAT1-1-1、MAT1-1-5、MAT1-2-1和MAT1-2-4,并存在与交配型转换相关的染色体倒位现象[7, 56]。其中,交配型MAT1-1-1、MAT1-1-5和MAT1-2-1基因缺陷型突变体能够形成产囊体但无法形成子囊盘,而MAT1-2-4基因突变则延缓菌核萌发并影响子囊盘形态及子囊孢子发育[55]。参与有性生殖调控的叉头框蛋白(SsFKH1)影响核盘菌发育及致病性[57];而另一蛋白(Ss-FoxE2)只与子囊盘的形成有关却不影响致病性[58]。此外,菌核的发育受到多种蛋白影响,其中特异性蛋白(Ssp1和Ssp2)参与菌核形成[59-60],黑色素合成蛋白(Scd1和Thr1)与抗逆性相关[61]。
核盘菌通过草酸调控寄主的氧化还原状态并引起侵染[10]。草酸合成相关的草酰乙酸乙酰水解酶(OAH)影响核盘菌形态建成和毒性[62],草酸脱羧酶(OxDC)影响附着孢发育及初侵染[63]。另外,活性氧(ROS)参与的氧化还原动态平衡是核盘菌引起病害的重要调控因子[10],如NADPH氧化酶(SsNox)影响草酸产生及菌核发育[64];过氧化氢酶(SCAT1)缺失引起菌丝分支及菌核发育异常且毒性降低[65];超氧化物歧化酶(SsSOD1)影响真菌毒性但与草酸及菌核的产生无关[66],而谷氨酰转肽酶(Ss-Ggt1)与菌核和附着孢发育及寄主互作相关[67]。
植物病原真菌通过分泌蛋白调控寄主反应建立侵染[11],利用单分子实时测序技术(Single molecule real-time sequencing technology)在基因组分析中鉴定了70个候选效应因子[11],转录组和蛋白质组水平也发现大量分泌蛋白(如效应因子)参与致病性或毒性[30, 68]。例如,分泌蛋白阿拉伯呋喃糖酶(Ssaxp)影响真菌毒性[69];富含半胱氨酸蛋白(SsCVNH)影响菌核发育及毒性[68],而富含半胱氨酸蛋白(SsSSVP1)却参与植物坏死反应[70];未知功能分泌蛋白(ssv263)与毒性有关[71],并用于植物病害诊断及流行监测[72];与附着孢形成相关的分泌蛋白(Ss-Caf1)也影响致病性及菌核发育[73];重排热点蛋白(Ss-Rhs1)基因沉默导致菌落形态异常及毒性降低[74];木聚糖蛋白酶(SsXyl1)基因缺失引起菌核异常并降低毒性[75]。
3 展望核盘菌作为一种毁灭性的植物病原真菌,在世界范围引起多种植物病害并造成严重经济损失。核盘菌引起的病害防控目前主要依赖化学杀菌剂,但随着抗药性、环境污染等问题出现及消费安全意识的提高,对经济、高效、环境友好的防控方案需求日益突出。了解植物病原真菌生物学基础,探索病害发生与发展规律,挖掘致病相关因子等研究为植物病害防控策略制定奠定基础。随着对复杂的生物系统的认知逐渐深入,生物学研究已经从分子水平发展到系统水平[76]。系统生物学(Systems biology)作为多学科交叉研究领域,建立基于组学数据的整合分析策略,揭示生物系统的组成结构与动态,代替仅关注单一分子或技术的传统研究方法[77]。因此,整合多组学数据信息并运用系统生物学研究方法论,更有助于全面认识植物病原真菌及其引起的植物病害系统。以基因组学、转录组学、蛋白组学、代谢组学为基础的组学技术在核盘菌研究中取得进展(图 1),为深层次研究核盘菌及其致病机制等提供了科学依据,但不同组学水平发掘的分子机理及调控网络等仍然有待梳理。随着核盘菌相关组学基础研究不断开展,加之生物技术的迅猛发展,核盘菌的系统生物学及分子调控互作机制等方面需要特别关注以下几个方面:扩展组学研究内涵,如利用脂类组学(Lipidomics)、糖组学(Glycomics)及互作组学(Interactomics)等;加强对真菌的基础与分子生物学方面的大数据分析,通过构建分子调控网络以发掘相关因子(如效应因子)及其功能;探索植物寄主与病原真菌间的识别与应答机理,了解二者互作及协同进化关系;完善植物及真菌的遗传操作技术,包括引入基因编辑CRISPR/Cas系统。通过上述方面的系统性研究,不仅有助于阐释核盘菌及其引起的病害系统研究,也为探索其他真菌及植物病害提供研究策略参考。
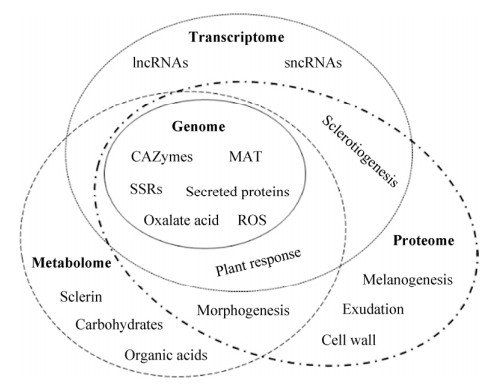 |
| 图 1 核盘菌的多组学分析体系 Fig. 1 Analysis system of multi-omics in Sclerotinia sclerotiorum. |
| 图选项 |
参考文献
| [1] | Bolton MD, Thomma BPHJ, Nelson BD. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen.Mol Plant Pathol, 2006, 7(1): 1–16. |
| [2] | Zhang WW, Li F, Nie L. Integrating multiple 'omics' analysis for microbial biology: application and methodologies.Microbiology, 2010, 156(2): 287–301.DOI: 10.1099/mic.0.034793-0 |
| [3] | Boland GJ, Hall R. Index of plant hosts of Sclerotinia sclerotiorum.Can J Plant Pathol, 1994, 16(2): 93–108.DOI: 10.1080/07060669409500766 |
| [4] | Silva RN. Perspectives in genomics the future of fungi in 'omics' era.Curr Genomics, 2016, 17(2): 82–84. |
| [5] | Grigoriev IV, Nikitin R, Haridas S, et al. MycoCosm portal: gearing up for 1000 fungal genomes.Nucleic Acids Res, 2014, 42(D1): D699–D704.DOI: 10.1093/nar/gkt1183 |
| [6] | Kan FL, Chen WX. Microbial genomic research and its impact on biological science.Acta Microbiol Sin, 2000, 40(3): 331–333.(in Chinese). 阚凤玲, 陈文新. 微生物基因组研究及其对生命科学的影响.微生物学报, 2000, 40(3): 331-333.DOI:10.3321/j.issn:0001-6209.2000.03.018 |
| [7] | Amselem J, Cuomo CA, van Kan JAL, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea.PLoS Genet, 2011, 7(8): e1002230.DOI: 10.1371/journal.pgen.1002230 |
| [8] | Freel KC, Friedrich A, Schacherer J. Mitochondrial genome evolution in yeasts: an all-encompassing view.FEMS Yeast Res, 2015, 15(4): fov023. |
| [9] | Heard S, Brown NA, Hammond-Kosack K. An interspecies comparative analysis of the predicted secretomes of the necrotrophic plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea.PLoS ONE, 2015, 10(6): e0130534.DOI: 10.1371/journal.pone.0130534 |
| [10] | Williams B, Kabbage M, Kim HJ, et al. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment.PLoS Pathog, 2011, 7(6): e1002107.DOI: 10.1371/journal.ppat.1002107 |
| [11] | Derbyshire M, Denton-Giles M, Hegedus D, et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens.Genome Biol Evol, 2017, 9(3): 593–618. |
| [12] | Bhadauria V, Popescu L, Zhao WS, et al. Fungal transcriptomics.Microbiol Res, 2007, 162(4): 285–298.DOI: 10.1016/j.micres.2007.06.006 |
| [13] | Adams MD, Kelley JM, Gocayne JD, et al. Complementary DNA sequencing: expressed sequence tags and human genome project.Science, 1991, 252(5013): 1651.DOI: 10.1126/science.2047873 |
| [14] | Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray.Science, 1995, 270(5235): 467.DOI: 10.1126/science.270.5235.467 |
| [15] | Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis.Genome Biol, 2016, 17(1): 13.DOI: 10.1186/s13059-016-0881-8 |
| [16] | Li RG, Rimmer R, Buchwaldt L, et al. Interaction of Sclerotinia sclerotiorum with a resistant Brassica napus cultivar: expressed sequence tag analysis identifies genes associated with fungal pathogenesis.Fungal Genet Biol, 2004, 41(8): 735–753.DOI: 10.1016/j.fgb.2004.03.001 |
| [17] | Sexton AC, Cozijnsen AJ, Keniry A, et al. Comparison of transcription of multiple genes at three developmental stages of the plant pathogen Sclerotinia sclerotiorum.FEMS Microbiol Lett, 2006, 258(1): 150–160.DOI: 10.1111/fml.2006.258.issue-1 |
| [18] | Oliveira MB, Junior ML, Grossi-de-Sá MF, et al. Exogenous application of methyl jasmonate induces a defense response and resistance against Sclerotinia sclerotiorum in dry bean plants.J Plant Physiol, 2015, 182: 13–22.DOI: 10.1016/j.jplph.2015.04.006 |
| [19] | Yang B, Srivastava S, Deyholos MK, et al. Transcriptional profiling of canola (Brassica napus L.) responses to the fungal pathogen Sclerotinia sclerotiorum.Plant Sci, 2007, 173(2): 156–171.DOI: 10.1016/j.plantsci.2007.04.012 |
| [20] | Calla B, Blahut-Beatty L, Koziol L, et al. Transcriptome analyses suggest a disturbance of iron homeostasis in soybean leaves during white mould disease establishment.Mol Plant Pathol, 2014, 15(6): 576–588.DOI: 10.1111/mpp.2014.15.issue-6 |
| [21] | Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity.EMBO Rep, 2001, 2(11): 986–991.DOI: 10.1093/embo-reports/kve230 |
| [22] | Joshi RK, Megha S, Rahman MH, et al. A global study of transcriptome dynamics in canola (Brassica napus L.) responsive to Sclerotinia sclerotiorum infection using RNA-Seq.Gene, 2016, 590(1): 57–67.DOI: 10.1016/j.gene.2016.06.003 |
| [23] | Zhou JH, Fu YP, Xie JT, et al. Identification of microRNA-like RNAs in a plant pathogenic fungus Sclerotinia sclerotiorum by high-throughput sequencing.Mol Genet Genomics, 2012, 287(4): 275–282.DOI: 10.1007/s00438-012-0678-8 |
| [24] | Wu J, Zhao Q, Yang QY, et al. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus.Sci Rep, 2016, 6: 19007.DOI: 10.1038/srep19007 |
| [25] | Girard IJ, Tong CB, Becker MG, et al. RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection.J Exp Bot, 2017, 68(18): 5079–5091.DOI: 10.1093/jxb/erx338 |
| [26] | Seifbarghi S, Borhan MH, Wei YD, et al. Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus.BMC Genomics, 2017, 18: 266.DOI: 10.1186/s12864-017-3642-5 |
| [27] | Zhang X, Fang AQ, Riley CP, et al. Multi-dimensional liquid chromatography in proteomics—a review.Anal Chim Acta, 2010, 664(2): 101–113.DOI: 10.1016/j.aca.2010.02.001 |
| [28] | Gonzalez-Fernandez R, Jorrin-Novo JV. Contribution of proteomics to the study of plant pathogenic fungi.J Proteome Res, 2012, 11(1): 3–16.DOI: 10.1021/pr200873p |
| [29] | Zieske LR. A perspective on the use of iTRAQTM reagent technology for protein complex and profiling studies.J Exp Bot, 2006, 57(7): 1501–1508.DOI: 10.1093/jxb/erj168 |
| [30] | Yajima W, Kav NNV. The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum.Proteomics, 2006, 6(22): 5995–6007.DOI: 10.1002/(ISSN)1615-9861 |
| [31] | Liu LZ, Free SJ. Characterization of the Sclerotinia sclerotiorum cell wall proteome.Mol Plant Pathol, 2016, 17(6): 985–995.DOI: 10.1111/mpp.12352 |
| [32] | Liang Y, Rahman MH, Strelkov SE, et al. Developmentally induced changes in the sclerotial proteome of Sclerotinia sclerotiorum.Fungal Biol, 2010, 114(8): 619–627.DOI: 10.1016/j.funbio.2010.05.003 |
| [33] | Liang Y, Strelkov SE, Kav NNV. The proteome of liquid sclerotial exudates from Sclerotinia sclerotiorum.J Proteome Res, 2010, 9(6): 3290–3298.DOI: 10.1021/pr900942w |
| [34] | Liang Y, Srivastava S, Rahman MH, et al. Proteome changes in leaves of Brassica napus L. as a result of Sclerotinia sclerotiorum challenge.J Agric Food Chem, 2008, 56(6): 1963–1976.DOI: 10.1021/jf073012d |
| [35] | Cao JY, Xu YP, Cai XZ. TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia sclerotiorum.J Proteomics, 2016, 143: 265–277.DOI: 10.1016/j.jprot.2016.03.006 |
| [36] | Wen L, Tan TL, Shu JB, et al. Using proteomic analysis to find the proteins involved in resistance against Sclerotinia sclerotiorum in adult Brassica napus.Eur J Plant Pathol, 2013, 137(3): 505–523.DOI: 10.1007/s10658-013-0262-z |
| [37] | Liang Y, Strelkov SE, Kav NNV. Oxalic acid-mediated stress responses in Brassica napus L.Proteomics, 2009, 9(11): 3156–3173.DOI: 10.1002/pmic.v9:11 |
| [38] | Wang JP, Xu YP, Zang XP, et al. Sclerotinia sclerotiorum virulence is affected by mycelial age via reduction in oxalate biosynthesis.J Integr Agric, 2016, 15(5): 1034–1045.DOI: 10.1016/S2095-3119(15)61199-6 |
| [39] | Nielsen J, Oliver S. The next wave in metabolome analysis.Trends Biotechnol, 2005, 23(11): 544–546.DOI: 10.1016/j.tibtech.2005.08.005 |
| [40] | Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics.Nat Rev Microbiol, 2005, 3(12): 937–947. |
| [41] | Penn CD, Daniel SL. Salicylate degradation by the fungal plant pathogen Sclerotinia sclerotiorum.Curr Microbiol, 2013, 67(2): 218–225.DOI: 10.1007/s00284-013-0349-y |
| [42] | Azevedo L, Chagas-Paula DA, Kim H, et al. White mold (Sclerotinia sclerotiorum), friend or foe: cytotoxic and mutagenic activities in vitro and in vivo.Food Res Int, 2016, 80: 27–35.DOI: 10.1016/j.foodres.2015.11.029 |
| [43] | Gárriz A, Dalmasso MC, Marina M, et al. Polyamine metabolism during the germination of Sclerotinia sclerotiorum ascospores and its relation with host infection.New Phytol, 2004, 161(3): 847–854. |
| [44] | Gárrriz A, Gonzalez ME, Marina M, et al. Polyamine metabolism during sclerotial development of Sclerotinia sclerotiorum.Mycol Res, 2008, 112(3): 414–422. |
| [45] | Pedras MSC, Ahiahonu PWK. Phytotoxin production and phytoalexin elicitation by the phytopathogenic fungus Sclerotinia sclerotiorum.J Chem Ecol, 2004, 30(11): 2163–2179. |
| [46] | Heuberger AL, Robison FM, Lyons SMA, et al. Evaluating plant immunity using mass spectrometry-based metabolomics workflows.Front Plant Sci, 2014, 5: 291. |
| [47] | Peluffo L, Lia V, Troglia C, et al. Metabolic profiles of sunflower genotypes with contrasting response to Sclerotinia sclerotiorum infection.Phytochemistry, 2010, 71(1): 70–80. |
| [48] | Robison FM, Turner M, Jahn CE, et al. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum.Plant Cell Environ, 2018, 41(9): 2141–2154. |
| [49] | Malen?i? D, Cveji? J, Tepav?evi? V, et al. Changes in L-phenylalanine ammonia-lyase activity and isoflavone phytoalexins accumulation in soybean seedlings infected with Sclerotinia sclerotiorum.Cent Eur J Biol, 2013, 8(9): 921–929. |
| [50] | Monazzah M, Soleimani MJ, Enferadi ST, et al. Effects of oxalic acid and culture filtrate of Sclerotinia sclerotiorum on metabolic changes in sunflower evaluated using FT-IR spectroscopy.J Gen Plant Pathol, 2018, 84(1): 2–11. |
| [51] | Wang XJ, Li JM, Zhu P. Effect of Sclerotinia sclerotiorum on lipid metabolism in Arabidopsis thaliana.J Plant Dis Prot, 2017, 124(5): 421–426. |
| [52] | Stotz HU, Sawada Y, Shimada Y, et al. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum.Plant J, 2011, 67(1): 81–93. |
| [53] | Weld RJ, Plummer KM, Carpenter MA, et al. Approaches to functional genomics in filamentous fungi.Cell Res, 2006, 16(1): 31–44. |
| [54] | Schmidt SM, Panstruga R. Pathogenomics of fungal plant parasites: what have we learnt about pathogenesis?.Curr Opin Plant Biol, 2011, 14(4): 392–399. |
| [55] | Doughan B, Rollins JA. Characterization of MAT gene functions in the life cycle of Sclerotinia sclerotiorum reveals a lineage-specific MAT gene functioning in apothecium morphogenesis.Fungal Biol, 2016, 120(9): 1105–1117. |
| [56] | Chitrampalam P, Inderbitzin P, Maruthachalam K, et al. The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation.PLoS ONE, 2013, 8(2): e56895. |
| [57] | Fan HD, Yu G, Liu YZ, et al. An atypical forkhead-containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum.Mol Plant Pathol, 2017, 18(7): 963–975. |
| [58] | Wang L, Liu YZ, Liu JL, et al. The Sclerotinia sclerotiorum FoxE2 gene is required for apothecial development.Phytopathology, 2016, 106(5): 484–490. |
| [59] | Li MY, Rollins JA. The development-specific protein (Ssp1) from Sclerotinia sclerotiorum is encoded by a novel gene expressed exclusively in sclerotium tissues.Mycologia, 2009, 101(1): 34–43. |
| [60] | Li MY, Rollins JA. The development-specific ssp1 and ssp2 genes of Sclerotinia sclerotiorum encode lectins with distinct yet compensatory regulation.Fungal Genet Biol, 2010, 47(6): 531–538. |
| [61] | Liang Y, Xiong W, Steinkellner S, et al. Deficiency of the melanin biosynthesis genes SCD1 and THR1 affects sclerotial development and vegetative growth, but not pathogenicity, in Sclerotinia sclerotiorum.Mol Plant Pathol, 2018, 19(6): 1444–1453. |
| [62] | Liang XF, Liberti D, Li MY, et al. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants.Mol Plant Pathol, 2015, 16(6): 559–571. |
| [63] | Liang XF, Moomaw EW, Rollins JA. Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function.Mol Plant Pathol, 2015, 16(8): 825–836. |
| [64] | Kim HJ, Chen CB, Kabbage M, et al. Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases.Appl Environ Microbiol, 2011, 77(21): 7721–7729. |
| [65] | Yarden O, Veluchamy S, Dickman MB, et al. Sclerotinia sclerotiorum catalase SCAT1 affects oxidative stress tolerance, regulates ergosterol levels and controls pathogenic development.Physiol Mol Plant Pathol, 2014, 85: 34–41. |
| [66] | Xu LS, Chen WD. Random T-DNA mutagenesis identifies a Cu/Zn superoxide dismutase gene as a virulence factor of Sclerotinia sclerotiorum.Mol Plant Microbe Interact, 2013, 26(4): 431–441.DOI: 10.1094/MPMI-07-12-0177-R |
| [67] | Li MY, Liang XF, Rollins JA. Sclerotinia sclerotiorum γ-glutamyl transpeptidase (Ss-Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria.Mol Plant Microbe Interact, 2012, 25(3): 412–420. |
| [68] | Lyu X, Shen CC, Fu YP, et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development.Sci Rep, 2015, 5: 15565. |
| [69] | Yajima W, Liang Y, Kav NNV. Gene disruption of an arabinofuranosidase/β-xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue.Mol Plant Microbe Interact, 2009, 22(7): 783–789.DOI: 10.1094/MPMI-22-7-0783 |
| [70] | Lyu X, Shen CC, Fu YP, et al. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants.PLoS Pathog, 2016, 12(2): e1005435. |
| [71] | Liang Y, Yajima W, Davis MR, et al. Disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on canola (Brassica napus).Can J Plant Pathol, 2013, 35(1): 46–55.DOI: 10.1080/07060661.2012.745904 |
| [72] | Ziesman BR, Turkington TK, Basu U, et al. A quantitative PCR system for measuring Sclerotinia sclerotiorum in canola (Brassica napus).Plant Dis, 2016, 100(5): 984–990. |
| [73] | Xiao XQ, Xie JT, Cheng JS, et al. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development.Mol Plant Microbe Interact, 2014, 27(1): 40–55. |
| [74] | Yu Y, Xiao JF, Zhu WJ, et al. Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum.Mol Plant Pathol, 2017, 18(8): 1052–1061. |
| [75] | Yu Y, Xiao JF, Du J, et al. Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of Sclerotinia sclerotiorum.Front Microbiol, 2016, 7: 1787. |
| [76] | Kitano H. Systems biology: a brief overview.Science, 2002, 295(5560): 1662–1664.DOI: 10.1126/science.1069492 |
| [77] | Durmu? S, ?ak?r T, ?zgür A, et al. A review on computational systems biology of pathogen-host interactions.Front Microbiol, 2015, 6: 235. |
