

湖北大学 生命科学学院 生物资源绿色转化湖北省协同创新中心,湖北 武汉 430062
收稿日期:2017-08-30;接收日期:2017-10-23 基金项目:国家重点研发计划(No. 2017YFD0501506)资助
摘要:玉米赤霉烯酮(Zearalenone,ZEN)及衍生物是一类主要由镰刀菌属的真菌产生的非甾体雌激素类真菌毒素,广泛存在于玉米、大麦、小麦和高粱等谷物饲料及其副产品中,严重危害牲畜及人类健康,迫切需要相关的技术对ZEN进行降解脱毒。传统的物理化学方法不能有效去除谷物中的毒素,并会破坏谷物的营养成分,影响食物口感,甚至造成二次污染,因此利用生物工程技术对ZEN及其衍生物进行脱毒是未来解决这一问题的主要方法。文中简要介绍了ZEN及衍生物和降解ZEN的微生物种类、降解特性,然后详细介绍了目前研究的ZEN降解酶种类、解析唯一的蛋白结构及其异源表达和应用情况,以期为通过分子酶工程和发酵工程等生物工程技术降低ZEN降解酶的成本提供指导,从而提高食品安全。
关键词:真菌毒素 玉米赤霉烯酮 玉米赤霉烯酮降解酶 异源表达 食品安全
Progress in bio-degradation of mycotoxin zearalenone
Li Shunyi, Yu Qiuxiang, Xiang La, Zhou Yuling, Zhang Guimin


Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, College of Life Sciences, Hubei University, Wuhan 430062, Hubei, China
Received: August 30, 2017; Accepted: October 23, 2017
Supported by: National Key Research and Development Program of China (No. 2017YFD0501506)
Corresponding author:Guimin Zhang. Tel: +86-27-88663882; E-mail: zhangguimin6@hotmail.com
Abstract: Zearalenone (ZEN) and its derivatives are non-steroidal estrogenic mycotoxins mainly produced by Fusarium species. They are widely distributed in grain feeds originated from maize, barley, wheat and sorghum, causing serious harm to animal and human health. Currently, there is a pressing need of an efficient technology for ZEN degradation and detoxification. Because traditional physical and chemical methods could not effectively detoxify ZEN in grains, and might also affect the grain nutrients and food taste, and even result in secondary pollution, the biological technologies are developed to detoxify ZEN and its derivatives. In this paper, we reviewed the structure of ZEN and its derivatives, the fungi and bacteria species with ability of degradation of ZEN. In addition, the characterization, protein sequences and conformation of currently identified ZEN degrading enzymes, the only solved ZHD structure from Clonostachys rose were analyzed and compared, and the enzymes heterologous expression and application were also reviewed. This review will provide reference for reducing the cost of ZEN degrading enzymes by biological technologies such as enzyme engineering and fermentation engineering.
Key words: mycotoxin zearalenone zearalenone degrading enzymes heterologous expression food safety
玉米赤霉烯酮(Zearalenone,ZEN)是一种主要由镰刀菌属Fusarium真菌产生的非甾体雌激素类真菌毒素,广泛存在于玉米、大麦、小麦和高粱等谷物及其副产品中。ZEN通过污染的谷物农副产品及饲料进入食物链,被动物吸收后,引起牲畜雌激素综合症状,导致牲畜体内雌激素过多,引起不孕不育、流产和死胎现象,并有很强的致癌性,严重危害牲畜及人类健康。有多篇文献对ZEN的合成、代谢、毒性、脱毒及控制等进行了综述[1-4]。研究ZEN的脱毒方法和技术是应对ZEN危害的主要措施。目前ZEN脱毒主要方法有物理法、化学法和生物法。物理法脱毒效率不高,而且破坏谷物营养成分,影响食品口感;化学法可以有效脱毒,但是如何从谷物食品中去掉脱毒用的化学成分很困难,容易造成二次污染。因此利用生物技术对ZEN及其衍生物进行降解和脱毒是未来解决这一问题的主要方法。生物技术脱毒主要是通过活菌降解和酶法降解两种途径。本文简要介绍了ZEN及衍生物的结构和降解ZEN的微生物种类、降解特性,然后详细介绍目前研究的ZEN降解酶的种类、蛋白结构及其异源表达和应用的情况。
1 ZEN及其衍生物ZEN首次由Stob等[5]于1962年从玉米中分离得到,1966年由Urry等[6]测定了化学结构,是一种酚的二羟基苯酸内脂结构化合物,分子式为C18H23O,平均分子量为318 Da,白色晶体,微溶于水,沸点为510.12 ℃,熔点为217.32 ℃。ZEN可以经过代谢转变为α-玉米赤霉烯醇(α-zearalenol,α-ZOL)、β-玉米赤霉烯醇(β-zearalenol,β-ZOL)、玉米赤霉酮(Zearalanone,ZAN)、α-玉米赤霉醇(α-zearalanol,a-ZAL)和β-玉米赤霉醇(β-zearalanol,β-ZAL)等几种衍生物(表 1)。ZEN及其衍生物与动物内源性雌激素β-雌二醇有类似的化学结构,故可以与动物细胞内β-雌二醇的受体结合而表现出毒性。研究表明,ZEN及其衍生物与鼠子宫胞浆的β-雌二醇受体的亲和力依次是α-ZAL﹥α-ZOL﹥β-ZAL﹥ZEN﹥β-ZOL[7],与雌激素受体的亲和力越强,则雌激素活性就越大,毒性也就越大。
表 1 玉米赤霉烯酮及其衍生物的结构Table 1 The structure of zearalenone and its derivatives
| Name | Structure |
| Zearalenone (ZEN) |  |
| α-Zearalenol (α-ZOL) |  |
| β-Zearalenol (β-ZOL) |  |
| Zearalanone (ZAN) |  |
| α-Zearalanol (α-ZAL) | 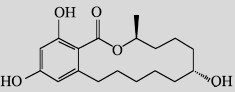 |
| β-Zearalanol (β-ZAL) |  |
| β-Estradiol |  |
表选项
ZEN通过霉变的玉米、小麦、高粱等食品和饲料被动物吸收,主要表现为亚急性和慢性毒性,急性毒性相对较低,小鼠口服LD50﹥20 000 mg/kg体重,大鼠口服LD50 > 10 000 mg/kg体重[8]。多国设定了食物和饲料中ZEN的最高限值。中国国家标准(GB 13078.2-2006)设定了饲料中ZEN的最大限量为500 μg/kg[9],而实际上我国谷物及其制品普遍存在高浓度的ZEN污染情况[10-12]。生物降解可以将ZEN高效转化为低毒或无毒的产物,环保安全,不会破坏谷物的营养物质,因此开发ZEN的脱毒技术具有重大意义。
2 活菌降解ZEN生物降解主要包括活菌降解法和酶降解法。活菌降解法是利用微生物对ZEN的转化从而降低其对动物的毒性。为了能降解ZEN,自20世纪80年代开始国内外的微生物学家就开始分离各种能降解ZEN的微生物,表 2为部分已经分离到的可以降解ZEN及其衍生物的真菌和细菌及其降解性质。活菌脱毒存在菌株培养时间长、降解活性低和后期菌株本身难以去除等问题,甚至有些菌株还是致病菌。因此近几年来,利用益生菌进行ZEN的降解研究越来越多,其中主要有酵母菌、芽孢杆菌和乳酸菌。
表 2 主要ZEN降解菌Table 2 Main strains with capability of ZEN degradation
| Microorganism | Capability of ZEN degradation | References | |
| Fungi | Aspergillus niger FS10 | 60.1% of 29 μg/mL ZEN was degraded in 48 h | [26] |
| Clonostachys catenulatum AN 169 | 100% degraded in 24 h | [27] | |
| Clonostachys rosea AN 154 | 50% degraded in 2 d, 80%–90% degraded in 8 d | [27] | |
| Clonostachys rose IFO 7063 | Cleavage ZEN to a non-estrogenic compound | [28] | |
| Gliocladium roseum | 80%–90% was degraded | [29] | |
| Mucor banieri | 30% was degraded | [30] | |
| Candida utilis CLY01 | 96.79% of 3.893 mg/L was degraded in 96 h | [16] | |
| Rhizopus stolonifer, Rhizopus oryzae, Rhizopus microsporus | 100% was degraded | [31] | |
| Rhodococcus pyridinivorans K404, K408 | 87.21% and 80.33% of 2 ppm was degraded | [32] | |
| Saccharomyces cerevisiae | Adsorb at a first state, and later 100% of 2.75 μg/mL was degraded in 48 h | [17-18] | |
| Streptomyces rimosus K145, K189 | 99% of 1 μg/mL was degraded in 5d | [33] | |
| Thamnidium elegans | 60% was degraded | [30] | |
| Trichoderma aggressivum AN 171 | 50% degraded in 6 d, 80%–90% degraded in 8 d | [27] | |
| Trichosporon mycotoxinivorans | 100% of 10 μg/mL was degraded in 24 h | [14-15] | |
| Bacteria | Acinetobacter sp. SM04 | 100% of 20 μg/mL was degraded in 12 h | [34] |
| Bacillus amyloliquefaciens | 93.92% of 10 μg/mL was degraded in 8 h | [20] | |
| Bacillus licheniformis 21-2 | 100% degraded from 5 μg/mL ZEN in 72 h | [35] | |
| Bacillus licheniformis CK1 | 95.8% of 2 ppm was degraded in 36 h | [36] | |
| Bacillus natto CICC2464 | Adsorb at a first state, and 100% of 20 μg/mL was degraded in 24 h | [37] | |
| Bacillus stratosphericus T246 | 100% of 15 μg/mL was degraded in 6 h | [38] | |
| Bacillus subtilis 168 | 81% of 20 μg/L was degraded in 24 h | [37] | |
| Bacillus subilis 373-2, 411-1 | 100% of 15 μg/mL was degraded in 8 and 6 h | [39] | |
| Bacillus subilis ANSB01G | 88.65% was degraded | [19] | |
| Bacillus subtilis BF-B-3 | 62.48% of 20 μg/mL was degraded in 4 d | [40] | |
| Bacillus pumilus T-420 | 100% of 15 μg/mL was degraded in 9 h | [38] | |
| Lactobacillus plantarum | 45% of 0.1 μg/mL was degraded in 48 h | [25] | |
| Lactobacillus plantarum MON03 | 78% of 50 μg/mL was degraded in 24 h | [41] | |
| Pseudomonas alcaliphila TH-C1 | 68% of 2 μg/mL was degraded in 72 h | [42] | |
| Pseudomonas citronelloli ASAG16 | 91.59% of 20 μg/mL was degraded in 6 d | [43] | |
| Pseudomonas otitidis TH-N1 | 59% of 2 μg/mL was degraded in 72 h | [44] | |
| Pseudomonas plecoglossicida TH-L1 | 57% of 2 μg/mL was degraded in 72 h | [42] | |
| Nate: Strains with underlines are probiotics listed in China National Standard of Food Additives (GB2760-2014)[25]. | |||
表选项
多种酵母菌具有降解ZEN的能力[13]。2004年Molnar等[14]筛选并通过18S rDNA鉴定了能降解ZEN的解毒毛孢酵母Trichosporon mycotoxinivoran,代谢产物中没有检测到α-ZOL或β-ZOL。该菌株也可以降解赭曲霉素A (Ochratoxin A)等其他真菌毒素,可以用于动物饲料的脱毒处理。2010年Vekiru等[15]继续对该酵母降解ZEN的机理进行了研究。他们用LC-MS/MS、LC-DAD和NMR等方法分离并鉴定了ZEN及其代谢产物。结果证明ZEN的内酯环被水解,体外试验显示产物不能与雌激素受体蛋白结合,所以没有毒性。吴晖等[16]报道了产朊假丝酵母菌株CLY01在添加ZEN的培养基中发酵,96 h后ZEN的去除率达96.79%,ZEN的浓度由起始的3.893 mg/L降为0.125 mg/L,该课题组还对ZEN降解酶进行过系列研究。Zhang等[17-18]报道酿酒酵母Saccharomyces cerevisiae (1×108 CFU/mL) 48 h能将发酵液中浓度为2.75 μg/mL的ZEN完全降解。芽孢杆菌是研究最多的能降解ZEN的益生菌。Lei等[19]从动物食道、发霉的食物和饲料及土壤等来源筛选得到36个菌株,其中有5个菌株能降解液体培养基中50%的ZEN,最强的一株对ZEN的降解率达到88.65%,经鉴定为枯草芽孢杆菌Bacillus subtilis ANSB01G。2016年张倩等[20]从135份赤霉病高发区土壤样品中筛选到一株解淀粉芽孢杆菌Fu2-3,表现出明显的ZEN降解效果,6 h内对ZEN的清除率达95%以上。乳杆菌能通过两种方式对ZEN脱毒。一是通过物理作用吸附ZEN,细胞实验表明,其吸附的浓度在0.79–62.82 μmol/L[21-22]。乳杆菌Lactobacillus plantarum应用到玉米饲料上,能降低玉米中ZEN的浓度,发酵4 d后,玉米中ZEN能降低68%–75%[23]。二是通过酶降解作用。Zhao等[24]报道从27种传统发酵食物中筛选到3株能降解ZEN的乳杆菌,48 h对溶液中的ZEN清除率约为45%。表 2所示为可以降解ZEN及其衍生物的真菌和细菌,其中有12种是《食品添加剂使用标准》(GB2760-2014)目录中的益生菌[25],其他具有降解活性的菌株以后也可能加入到益生菌目录中。从表 2可以看出芽孢杆菌是报道最多、对ZEN有较好降解效果的益生菌,枯草芽孢杆菌、地衣芽孢杆菌等益生菌更是广泛用于畜牧水产饲料添加剂,如果能增强其降解ZEN的功能,将具有重要应用价值。
原始菌株具有不同的ZEN降解能力,但是因为菌株培养时间长、降解活性低等问题,在实际脱毒中应用尚需要对菌种进行选育。因此研究ZEN降解酶的结构和性质及降解的机制,具有重要的意义,开发ZEN降解酶是未来ZEN降解的发展趋势。
3 ZEN降解酶的发现上述能对ZEN脱毒的微生物主要方式是微生物能产生酶水解或氧化ZEN,从而破坏其结构并转变为无毒的小分子代谢物。有多种ZEN脱毒关键酶的研究和报道[45]。2002年Takahashi-Ando等[46]发现粉红粘帚霉Clonostachys rosea IFO 7063能将ZEN降解为低毒性的产物,他们从含ZEN的培养基培养的C. rosea IFO 7063中提取了一种碱性水解酶ZHD101,测定了部分氨基酸残基序列,根据氨基酸序列设计引物克隆了该酶的基因zhd101,并在裂殖酵母Schizosaccharomyces pombe和大肠杆菌Escherichia coli中得到表达,表达的产物具有酶活性,能水解ZEN。这是首次鉴定并克隆ZHD101酶基因的报道,对后续ZEN的生物法降解具有重要指导意义。2005年Utermark等[47]通过基因zhd101突变的方法证实ZHD101能降解ZEN。
除了ZHD101,ZEN降解酶的报道并不多,利用ZEN作为关键词搜索UniProt和PDB数据库,选择UniProt数据库及文献报道的19种酶进行了分析,图 1为根据这些酶的序列比对构建的系统进化树。其中第一分支为同源性最高的,有13种ZEN降解酶,分别为来源于C. rosea的Q8NKB0[46] (PDB ID 2WZL)、A0A023IYG7[47]、A0A023IYG2[48]、A0A023IYH9[48]、W0LXP2[27]和AN 154[27],链孢粘帚菌Clonostachys catenulatum AN 169的AN 169[27],淡色生赤壳菌Bionectria ochroleuca的A0A0N9XBU7 (Uniprot)、C9EIK3 (Uniprot)、G9C2T3 (Uniprot)和W0M193[27],侵占木霉Trichoderma aggressivum的W0M1M6[27]和AN 171[27]。这一组ZEN降解酶由264个氨基酸残基组成,分子量约为28 kDa,氨基酸序列同源性大于95.8%,其中W0LXP2和AN 169、W0M1M6和AN 154具有一致的序列,属于ZHD101的高度同源蛋白。
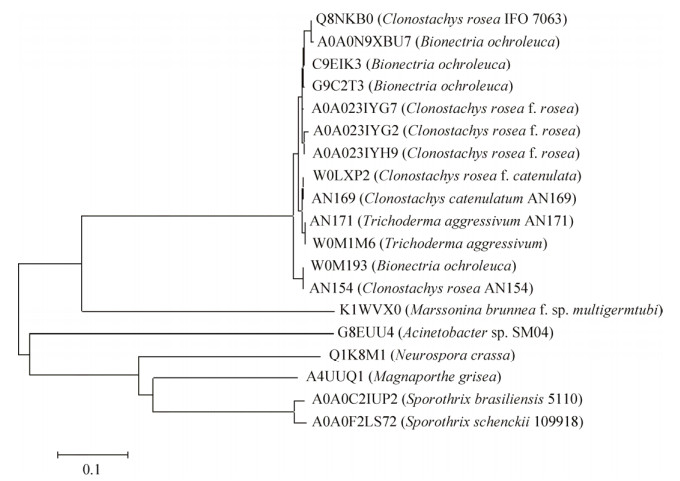 |
| 图 1 ZEN降解酶的系统发育树(酶的代号为UniProt或PDB数据库中的ID) Figure 1 Neighbor-joining phylogenetic tree of ZEN degradation enzyme. The enzyme codes are the ID in Uniprot or PDB databases. |
| 图选项 |
进化树分支另一端不动杆菌Acinetobacter sp. SM04的G8EUU4是另外一种ZEN降解酶。2011年Yu等[34]筛选到该菌可以对ZEN脱毒,2012年,Yu等[49]又鉴定出其对ZEN有脱毒功能的是过氧化物酶(Peroxiredoxin,EC 1.11.1.15),他们克隆了过氧化物酶基因并在大肠杆菌中成功表达。毒性试验表明,过氧化物酶在过氧化氢存在的条件下具有ZEN脱毒效果,但是过氧化氢的存在可能会影响该酶在食品和饲料中的应用。
其他来源的ZEN降解酶有稻瘟病菌Magnaporthe grisea的A4UUQ1 (Uniprot)、杨树黑斑病菌Marssonina brunnea f. sp. multigermtubi (strain MB_m1)的K1WVX0[50]、粗糙脉孢菌Neurospora crassa (strain ATCC 24698/ 74-OR23-1A/CBS 708.71/DSM 1257/FGSC 987)的Q1K8M1[51]、巴西孢子丝菌Sporothrix brasiliensis 5110的A0A0C2IUP2、申克孢子丝菌Sporothrix schenckii 1099-18的A0A0F2LS72[52]。这些酶彼此差别较大,与ZHD101相比也有较大的差别,同源性在11%–32%之间,其中与ZHD101同源性最高的是M. brunneai的K1WVX0[50],同源性为32%。以上5种酶是通过对基因组进行基因功能注释得到的疑似ZEN降解酶,并没有对具体的基因进行过克隆表达和功能研究。这些微生物的ZEN降解酶有待深入研究,以探索是否可以发现更有效的ZEN降解酶。
4 酶蛋白的结构及催化作用分子机理4.1 ZEN降解酶的结构模拟及分类2014年Popiel等[27]分析了ZHD101及同源性很高的C. catenulatum AN 169、C. rosea AN 154和T. aggressivum AN 171的ZHD酶氨基酸序列,发现它们与α/β水解酶折叠家族的3-氧代己二酸烯酸内酯酶(PDB ID 2XUA)和过氧化物α/β水解酶Lpx1 (PDB ID 2Y6U)的氨基酸序列有较大的同源性,于是以2XUA和2Y6U为模板模拟了3种ZHD酶的构象,结果显示它们都具有α/β水解酶折叠家族(ab-hydrolase)的空间构象特征。α/β水解酶折叠家族是一种广泛存在的结构类型,一般由5个以上的β-折叠组成的内核结构域和覆盖内核上的α-螺旋组成的帽子结构域组成,并有一个由亲核氨基酸、组氨酸和另外一个氨基酸组成催化三联体[53]。ZHD属于Abhydrolase_6,目前该家族共有3 819个基因,有53个蛋白质结构的报道(http://bioweb.ensam.inra.fr/esther)[54]。
4.2 ZEN降解酶的晶体结构ZEN降解酶的晶体结构解析首先由中国科学院天津工业生物技术研究所的郭瑞庭团队[55]完成。郭等用X-射线衍射的方法首次测定了来源于C. rosea的ZEN水解酶ZHD (PDB ID 3WZL)和ZHD-ZEN复合物(PDB ID 3WZM)的晶体结构,图 2为PyMOL显示的ZHD与底物ZEN复合物的晶体结构图。在此基础上,他们又测定了ZHD的单突变(S102A)和双突变体(S102A/V153H)及其与ZEN的复合体的结构[56]。结构分析显示该酶具有以下特点。1)酶蛋白由两个结构域组成:一个是α/β水解酶折叠结构域,由8段β-折叠组成的内核和两侧包围的7段α-螺旋组成;另一个是覆盖于其上的帽子结构域,由4段α-螺旋组成,图 3为ZEN降解酶与α/β水解酶折叠蛋白的空间结构模块示意图。这个结果与上述2014年Popeil等[27]模拟的构象相似,表明模拟的结果是正确的。2)酶与底物结合的位点位于两个结构域之间的空穴,底物ZEN通过3个氢键和几个非极性键嵌合到这个空穴中。Trp183是ZEN和酶结合的重要位点,当Trp183突变为Ala、His或Phe时,酶活性丧失。Leu33、Val153、Met154、Val158、Leu135、Phe221、Pro128、Ile191和Pro192等氨基酸的侧链也与底物直接接触,是酶与底物结合的保守位点。3)酶的活性中心为Ser102-His242-Glu126组成的催化三联体(图 2),Ser102羟基的O原子作为亲核基团攻击底物ZEN内酯环上的羰基C原子,从而引发水解反应。郭的团队还对该酶进行了一系列的氨基酸定点突变研究,突变体V153H蛋白对α-ZOL水解的比活力增加了3.7倍,Km增加了2.7倍,但kcat/Km增加了5.2倍,结构分析显示可能是因为V153H突变使得催化三联体中的H242获得了更大的空间。这些结构和系列突变酶的研究揭示了酶的催化反应机理,为提高ZHD的催化能力和推进ZHD的应用提供了重要的理论基础。
 |
| 图 2 PyMOL显示的ZHD与底物ZEN复合物的晶体结构(PDB ID 3WZM) Figure 2 Structure of ZHD and ZEN complex (PDB ID 3WZM) using program PyMOL. |
| 图选项 |
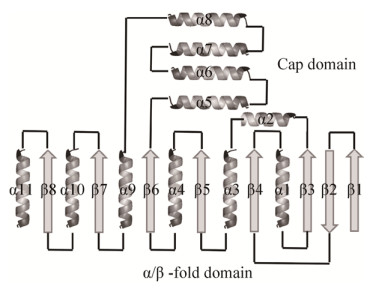 |
| 图 3 ZHD及α/β水解酶折叠家族的拓扑结构示意图 Figure 3 Toplogy diagrams of ZHD and α/β-hydrolase fold enzymes. |
| 图选项 |
5 ZEN降解酶的异源表达由于天然ZEN降解酶的产量及活性较低,对这些酶的异源高效表达成了ZEN高效降解的希望。我们对ZEN降解酶的种类来源及异源表达的研究汇总见表 3。异源表达研究最多的是C. rosea的zhd101基因,目前该基因已经在大肠杆菌、酿酒酵母、毕赤酵母和水稻中得到表达。2002年Takahashi-Ando等[47]在E. coli中成功表达了zhd101基因,但在S. pombe中却没有表达。2004年,Takahashi-Ando等[57]为了更直观地检测ZHD101解毒活性,在E. coli和S. cerevisiae中表达了ZHD101和绿色荧光蛋白的融合蛋白,重组大肠杆菌表现出较强的ZEN降解酶活性,但重组酿酒酵母ZEN降解酶活性则较弱,培养基中添加2 μg/mL ZEN或ZOL,重组大肠杆菌1 h能将其完全降解,而重组酿酒酵母4 d只能降解其中的75%。随后Takahashi-Ando[58]对zhd101基因进行了密码子优化,研究转基因酿酒酵母对ZEN的降解能力,结果表明该基因的表达水平增加了约20倍,28 ℃ 48 h或37 ℃ 8 h就能完全降解培养基中2 μg/mL的ZEN,且降解产物中没有检测到β-ZOL。2005年,Higa-Nishiyama等[59]将zhd101基因与增强的绿色荧光蛋白基因融合,融合基因在水稻叶片中成功表达,能有效降解ZEN毒素,且在种子内也检测到ZEN降解活性。国内的程波财等[60]从粉红螺旋聚孢霉中克隆到ZEN-jjm基因并在E. coli和毕赤酵母GS115中得到异源表达,大肠杆菌破菌上清液在3 h内能完全降解浓度为1 μg/mL的ZEN等[60],毕赤酵母发酵上清液需要9 h才能降解浓度为1 μg/mL的ZEN[61]。刘海燕等[62]从粉红粘帚霉菌中克隆到zlhy-6基因,并在P. pastoris GS115中得到表达,发酵上清4 h能降解浓度为1.6 μg/mL的ZEN。与zhd101基因相比对,ZEN-jjm基因和zlhy-6基因的同源性高达98%。2015年,王义春等[63]用多拷贝技术构建了zlhy-6基因表达工程菌并在巴斯德毕赤酵母中实现分泌表达,发酵液中的酶活力达到10 U/mL,1 h内对浓度为20 μg/mL的ZEN降解率达99%以上。
表 3 主要ZEN降解酶来源、异源表达及特征Table 3 The origin, heterologous expression and properties of main ZEN hydrolases
| Name | Microorganism | Host cell | Properties | References |
| Zhd101 | Clonostachys rosea | E. coli BL21 (DE3) | 100% of 2 μg/mL ZEN was detoxified in 1 h | [62] |
| S. cerevisiae | 75% of ZEN was detoxified in 4 d | [62] | ||
| P. pastoris GS115 | 90% of 10 μg/mL ZEN was degraded in 12 min by recombinant ZHD | [61] | ||
| L. reuteri | ZEN concentration decreased from 4.5 mg/L to 0.03 mg/L in 14 h | [63] | ||
| ZEN-jjm | Gliocladium roseum | E. coli BL21 (DE3) | 100% of 1 μg/mL ZEN was detoxified in 3 h | [57] |
| P. pastoris GS115 | 100% of 1 μg/mL ZEN was detoxified in 9 h | [58] | ||
| ZLHY-6 | Gliocladium roseum (31535) | P. pastoris GS115 | 100% of 1.6 μg/mL ZEN was detoxified in 4 h | [59] |
| Zearalenone lactonase | Pseudomonas putida ZEA-1 | E. coli BL21 (DE3) | 100% of 100 μg/mL ZEN was detoxified in 12 h | [64] |
| Peroxiredoxin | Acinetobacter sp. SM04 | E. coli BL21 (DE3) | 90% of 1 000 μg/mL ZEN was detoxified in 6 h with additional of 0.09% of H2O2 | [46] |
| S. cerevisiae INVSc1 | 57.5% of 20 μg/mL ZEN was detoxified in 12 h | [65] |
表选项
为了进一步探索ZHD101的应用,我们对zhd101基因(GenBank No. AB076037.1)进行了密码子优化(优化的基因GenBank No. KX229746)并首次在毕赤酵母中实现了高效表达[61]。结果表明含有3拷贝的转化子酶蛋白分泌量最高,表达上清可以有效降解ZEN及其衍生物α-ZOL和β-ZOL,10 μg/mL的ZEN经过15 min就降解到3.66 μg/mL,30 min内被完全降解。5 L发酵罐高密度发酵后的酶活力达到150.1 U/mL,是摇瓶发酵酶活力的6.7倍。在酿酒酵母发酵前用ZHD酶处理麦芽汁底物,结果表明,90%的ZEN在ZHD101酶加入后12 min内就被降解了,该研究为ZHD酶的应用提供了一种新的途径。
6 ZEN生物降解的展望尽管已经有多篇文章对ZEN生物降解进行了研究,取得了一定的成果,但离实际应用还存在一定的距离。为了实现对ZEN的高效生物脱毒,我们认为需要从以下几方面深入研究。1) ZEN脱毒酶基因资源的深度挖掘。目前真正报道有ZEN降解活性的酶就只有ZHD101及其同源性95%以上的同源物,因此克隆新的ZEN降解酶基因是未来的一个发展方向。2)酶的蛋白质工程。蛋白质工程是提高酶活性改善酶性能的重要手段,目前ZHD101的结构已经解析,相信已经有很多课题组在基于结构的基础上在对该酶进行改造,提高酶比活力和稳定性。3)酶的高效表达和制备技术。目前ZEN降解酶通过异源表达系统表达的最高酶活报道是150.1 U/mL,但是蛋白的表达量很低,还有很大的提升空间,可以利用基因工程和发酵工程技术进一步提高酶的产量,降低应用成本。如能得到更高效降解ZEN的菌株或酶,为ZEN的生物降解提供可行的技术和工艺,将对饲料安全和食品安全具有重要意义。
参考文献
| [1] | Zinedine A, Soriano JM, Moltó JC, et al. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: an oestrogenic mycotoxin.Food Chem Toxicol, 2007, 45(1): 1–18.DOI: 10.1016/j.fct.2006.07.030 |
| [2] | Shi J, Wang Y, Ju X, et al. Research advancement on biodegradation of zearalenone.J Chin Cereal Oil Ass, 2013, 28(6): 111–114.(in Chinese). 史競, 汪洋, 鞠星, 等. 玉米赤霉烯酮生物降解研究进展.中国粮油学报, 2013, 28(6): 111-114. |
| [3] | Ji C. Biodegradation of zearalenone in feeds.Acta Zoonutr Sin, 2014, 26(10): 2949–2955.(in Chinese). 计成. 饲料中玉米赤霉烯酮的生物降解.动物营养学报, 2014, 26(10): 2949-2955.DOI:10.3969/j.issn.1006-267x.2014.10.005 |
| [4] | Dellafiora L, Dall'Asta C. Forthcoming challenges in mycotoxins toxicology research for safer food—a need for multi-omics approach.Toxins, 2017, 9(1): 18.DOI: 10.3390/toxins9010018 |
| [5] | Stob M, Baldwin R, Tuite J, et al. Isolation of an anabolic, uterotrophic compound from corn infected with Gibberella zeae.Nature, 1962, 196(4861): 1318–1318. |
| [6] | Urry W, Wehrmeister H, Hodge E, et al. The structure of zearalenone.Tetrahedron Lett, 1966, 7(27): 3109–3114.DOI: 10.1016/S0040-4039(01)99923-X |
| [7] | Tashiro F, Kawabata Y, Naoi M, et al. eds. Zearalenone-estrogen receptor interaction and RNA synthesis in rat uterus. Medical Mycology, ed. Preusser HJ. Vol. Suppl. 8, Gustav Fisher Verlag: New York. 1980: 311–320. |
| [8] | Fifty-third meeting of the Joint FAO/WHO Expert Committee on Food Additives (JECFA), Safety evaluation of certain food additives and contaminants: WHO food additives series 44. Geneva, 2000. |
| [9] | Hygienical standard for feeds-Toleration of ochratoxin A and zearalenone in feeds. China National Standard GB 13078.2-2006 (in Chinese). 饲料卫生标准, 饲料中赭曲霉毒素A和玉米赤霉烯酮的允许量, 中华人民共和国国家标准GB 13078. 2-2006. |
| [10] | Wang W, Zhu JH, Shao B, et al. Probabilistic assessment of dietary exposure to both deoxynivalenol and zearalenone from cereal-based products in Chinese populations.Chin J Prevent Med, 2015, 49(3): 223–227.(in Chinese). 王伟, 朱江辉, 邵兵, 等. 中国居民小麦粉和玉米制品中脱氧雪腐镰刀菌烯醇和玉米赤霉烯酮暴露水平评估.中华预防医学杂志, 2015, 49(3): 223-227. |
| [11] | Xie D, Deng CL, Zhao YF, et al. Investigation of the contamination status of zearalenone and deoxynivalenol in edible oil.Food Safe Qual Detec Technol, 2016, 7(5): 2105–2113.(in Chinese). 谢丹, 邓春丽, 赵云峰, 等. 北京地区部分市售食用植物油中玉米赤霉烯酮和脱氧雪腐镰刀菌烯醇的污染状况分析.食品安全质量检测学报, 2016, 7(5): 2105-2113. |
| [12] | Ma CG, Wang YD. Research progress on pollution status and toxicity of zearalenone.J Henan Univ Technol: Nat Sci Ed, 2017, 38(1): 122–128.(in Chinese). 马传国, 王英丹. 玉米赤霉烯酮污染状况及毒性的研究进展.河南工业大学学报:自然科学版, 2017, 38(1): 122-128. |
| [13] | Dong MJ, Yang QY, Sun W, et al. A review on the control of zearalenone with antagonistic yeast.Food Sci (Beijing), 2016, 37(1): 230–234.(in Chinese). 董曼佳, 杨其亚, 孙伟, 等. 拮抗酵母菌控制玉米赤霉烯酮的研究进展.食品科学, 2016, 37(1): 230-234.DOI:10.7506/spkx1002-6630-201601040 |
| [14] | Molnar O, Schatzmayr G, Fuchs E, et al. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins.Syst Appl Microbiol, 2004, 27(6): 661–671.DOI: 10.1078/0723202042369947 |
| [15] | Vekiru E, Hametner C, Mitterbauer R, et al. Cleavage of zearalenone by Trichosporon mycotoxinivorans to a novel nonestrogenic metabolite.Appl Environ Microbiol, 2010, 76(7): 2353–2359.DOI: 10.1128/AEM.01438-09 |
| [16] | Wu H, Xian JW, Liu DM. Preliminary study on a zearalenone elimination yeast.Science and Technol Food Ind, 2009, 30(12): 215–216.(in Chinese). 吴晖, 冼嘉雯, 刘冬梅. 一株去除玉米赤霉烯酮酵母的初步研究.食品工业科技, 2009, 30(12): 215-216. |
| [17] | Keller L, Abrunhosa L, Keller K, et al. Zearalenone and its derivatives alpha-zearalenol and beta-zearalenol decontamination by Saccharomyces cerevisiae strains isolated from bovine forage.Toxins (Basel), 2015, 7(8): 3297–3308.DOI: 10.3390/toxins7083297 |
| [18] | Zhang H, Dong M, Yang Q, et al. Biodegradation of zearalenone by Saccharomyces cerevisiae: possible involvement of ZEN responsive proteins of the yeast.J Proteomics, 2016, 143: 416–423.DOI: 10.1016/j.jprot.2016.04.017 |
| [19] | Lei YP, Zhao LH, Ma QG, et al. Degradation of zearalenone in swine feed and feed ingredients by Bacillus subtilis ANSB01G.World Mycotoxin J, 2014, 7(2): 143–151.DOI: 10.3920/WMJ2013.1623 |
| [20] | Zhang Q, Xiong Q, Zhao C, et al. Preliminary study on screen of detoxified strain of zearalenone and mechanism of detoxification.Sci Technol Cereals, Oils Foods, 2016, 24(6): 76–81.(in Chinese). 张倩, 熊犍, 赵晨, 等. 玉米赤霉烯酮脱毒菌株的筛选及脱毒机理初探.粮油食品科技, 2016, 24(6): 76-81. |
| [21] | El-Nezami H, Polychronaki N, Salminen S, et al. Binding rather than metabolism may explain the interaction of two food-grade Lactobacillus strains with zearalenone and its derivative (')alpha-earalenol.Appl Environ Microbiol, 2002, 68(7): 3545–3549.DOI: 10.1128/AEM.68.7.3545-3549.2002 |
| [22] | El-Nezami H, Polychronaki N, Lee YK, et al. Chemical moieties and interactions involved in the binding of zearalenone to the surface of Lactobacillus rhamnosus strains GG.J Agric Food Chem, 2004, 52(14): 4577–4581.DOI: 10.1021/jf049924m |
| [23] | Mokoena MP, Chelule PK, Gqaleni N. Reduction of fumonisin B1 and zearalenone by lactic acid bacteria in fermented maize meal.J Food Prot, 2005, 68(10): 2095–2099.DOI: 10.4315/0362-028X-68.10.2095 |
| [24] | Zhao L, Jin H, Lan J, et al. Detoxification of zearalenone by three strains of lactobacillus plantarum from fermented food in vitro.Food Control, 2015, 54: 158–164.DOI: 10.1016/j.foodcont.2015.02.003 |
| [25] | Standard for food addivitives. China National Standard GB 2760-2014. 食品添加剂使用标准. 中华人民共和国国家标准GB 2760-2014. |
| [26] | Sun X, He X, siyu Xue K, et al. Biological detoxification of zearalenone by Aspergillus niger strain FS10.Food Chem Toxicol, 2014, 72(3): 76–82. |
| [27] | Popiel D, Koczyk G, Dawidziuk A, et al. Zearalenone lactonohydrolase activity in Hypocreales and its evolutionary relationships within the epoxide hydrolase subset of a/b-hydrolases.BMC Microbiol, 2014, 14(1): 82.DOI: 10.1186/1471-2180-14-82 |
| [28] | Kakeya H, Takahashi-Ando N, Kimura M, et al. Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of Clonostachys sp.Biosci Biotechnol Biochem, 2002, 66(12): 2723–2726.DOI: 10.1271/bbb.66.2723 |
| [29] | El-Sharkawy S, Abul-Hajj YJ. Microbial cleavage of zearalenone.Xenobiotica, 1988, 18(4): 365–371.DOI: 10.3109/00498258809041672 |
| [30] | El-Sharkawy S, Abul-Hajj Y. Microbial transformation of zearalenone, I. Formation of zearalenone-4-O-β-glucoside.J Nat Prod, 1987, 50(3): 520–521.DOI: 10.1021/np50051a038 |
| [31] | Varga J, Péteri Z, Tábori K, et al. Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates.Int J Food Microbiol, 2005, 99(3): 321–328.DOI: 10.1016/j.ijfoodmicro.2004.10.034 |
| [32] | Cserháti M, Kriszt B, Krifaton C, et al. Mycotoxin-degradation profile of Rhodococcus strains.Int J Food Microbiol, 2013, 166(1): 176–185.DOI: 10.1016/j.ijfoodmicro.2013.06.002 |
| [33] | Harkai P, Szabó I, Cserháti M, et al. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection.Int Biodeter Biodegr, 2016, 108: 48–56.DOI: 10.1016/j.ibiod.2015.12.007 |
| [34] | Yu Y, Qiu L, Wu H, et al. Degradation of zearalenone by the extracellular extracts of Acinetobacter sp. SM04 liquid cultures.Biodegradation, 2011, 22(3): 613–622.DOI: 10.1007/s10532-010-9435-z |
| [35] | Liu YX. Screening and identification of zearalenone-degrading strain and optimization of ZEN-degrading conditions[D]. Guangzhou: South China University of Technology, 2011 (in Chinese). 刘玉霞. 玉米赤霉烯酮降解菌的筛选、鉴定及脱毒条件研究[D]. 广州: 华南理工大学, 2011. |
| [36] | Yi PJ, Pai CK, Liu JR. Isolation and characterization of a Bacillus licheniformis strain capable of degrading zearalenone.World J Microbiol Biotechnol, 2011, 27(5): 1035–1043.DOI: 10.1007/s11274-010-0548-7 |
| [37] | Tinyiro SE, Wokadala C, Xu D, et al. Adsorption and degradation of zearalenone by Bacillus strains.Folia Microbiol, 2011, 56(4): 321.DOI: 10.1007/s12223-011-0047-8 |
| [38] | Ge CC, Xiong Q, Zhao C, et al. Screening of Bacillus being able to degrade zearalenone.Sci Technol Cereals, Oils Foods, 2015, 23(3): 90–94.(in Chinese). 葛婵婵, 熊犍, 赵晨, 等. 降解玉米赤霉烯酮的芽孢杆菌筛选.粮油食品科技, 2015, 23(3): 90-94. |
| [39] | Ge CC, Xiong Q, Zhao C, et al. Screening and identification of degradation zearalenone Bacillus.Sci Technol Cereals, Oils Foods, 2016, 24(3): 86–91.(in Chinese). 葛婵婵, 熊犍, 赵晨, 等. 降解玉米赤霉烯酮芽孢杆菌的筛选和鉴定.粮油食品科技, 2016, 24(3): 86-91. |
| [40] | Sun ZX, Xian YY, Sun WH, et al. Isolation, identification of a zearalenone degradated strain and the properties of its producing enzymes.Microbiol China, 2014, 41(5): 909–915.(in Chinese). 孙志轩, 冼钰茵, 孙纬华, 等. 分解玉米赤霉烯酮菌株的分离、鉴定及其产酶特征.微生物学通报, 2014, 41(5): 909-915. |
| [41] | Abbes S, Ben Salah-Abbes J, Sharafi H, et al. Interaction of Lactobacillus plantarum MON03 with Tunisian montmorillonite clay and ability of the composite to immobilize zearalenone in vitro and counteract immunotoxicity in vivo.Immunopharmacol Immunotoxicol, 2012, 34(6): 944–950.DOI: 10.3109/08923973.2012.674139 |
| [42] | Tan H, Hu Y, He J, et al. Zearalenone degradation by two Pseudomonas strains from soil.Mycotoxin Res, 2014, 30(4): 191–196.DOI: 10.1007/s12550-014-0199-x |
| [43] | Wang GB, Wu SL, Lin AJ, et al. Isolation and degradation characteristics of zearalenone-degrading bacterium.J Chin Cereals Oils Association, 2016, 31(1): 84–89.(in Chinese). 王国兵, 伍松陵, 林爱军, 等. 玉米赤霉烯酮降解菌的分离及降解特性研究.中国粮油学报, 2016, 31(1): 84-89. |
| [44] | Tan H, Zhang Z, Hu Y, et al. Isolation and characterization of Pseudomonas otitidis TH-N1 capable of degrading zearalenone.Food Control, 2015, 47: 285–290.DOI: 10.1016/j.foodcont.2014.07.013 |
| [45] | Tang YQ, Zhong F, Chen Y, et al. Review on biological detoxification of zearalenone and mechanism of key enzyme.Modern Food Sci Technol, 2013, 29(7): 1742–1746.(in Chinese). 唐语谦, 钟凤, 陈艺, 等. 玉米赤霉烯酮生物脱毒及关键酶作用机理的研究进展.现代食品科技, 2013, 29(7): 1742-1746. |
| [46] | Takahashi-Ando N, Kimura M, Kakeya H, et al. A novel lactonohydrolase responsible for the detoxification of zearalenone: enzyme purification and gene cloning.Biochem J, 2002, 365(Pt 1): 1–6. |
| [47] | Utermark J, Karlovsky P. Role of zearalenone lactonase in protection of Gliocladium roseum from fungitoxic effects of the mycotoxin zearalenone.Appl Environ Microb, 2007, 73(2): 637–642.DOI: 10.1128/AEM.01440-06 |
| [48] | Kosawang C, Karlsson M, Vél z H, et al. Zearalenone detoxification by zearalenone hydrolase is important for the antagonistic ability of Clonostachys rosea against mycotoxigenic Fusarium graminearum.Fungal Biology, 2014, 118(4): 364–373.DOI: 10.1016/j.funbio.2014.01.005 |
| [49] | Yu Y, Wu H, Tang Y, et al. Cloning, expression of a peroxiredoxin gene from Acinetobacter sp. SM04 and characterization of its recombinant protein for zearalenone detoxification.Microbiol Res, 2012, 167(3): 121–126.DOI: 10.1016/j.micres.2011.07.004 |
| [50] | Zhu S, Cao YZ, Jiang C, et al. Sequencing the genome of Marssonina brunnea reveals fungus-poplar co-evolution.BMC Genomics, 2012, 13(1): 382.DOI: 10.1186/1471-2164-13-382 |
| [51] | Galagan JE, Calvo SE, Borkovich KA, et al. The genome sequence of the filamentous fungus Neurospora crassa.Nature, 2003, 422(6934): 859.DOI: 10.1038/nature01554 |
| [52] | Teixeira MM, de Almeida LG, Kubitschek-Barreira P, et al. Comparative genomics of the major fungal agents of human and animal sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis.BMC Genomics, 2014, 15(1): 943.DOI: 10.1186/1471-2164-15-943 |
| [53] | Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing.Curr Opin Struct Biol, 1999, 9(6): 732–737.DOI: 10.1016/S0959-440X(99)00037-8 |
| [54] | Lenfant N, Hotelier T, Bourne Y, et al. Proteins with an alpha/beta hydrolase fold: Relationships between subfamilies in an ever-growing superfamily.Chem Biol Interact, 2013, 203(1): 266–268.DOI: 10.1016/j.cbi.2012.09.003 |
| [55] | Peng W, Ko TP, Yang Y, et al. Crystal structure and substrate-binding mode of the mycoestrogen-detoxifying lactonase ZHD from Clonostachys rosea.Rsc Advances, 2014, 4(107): 62321–62325.DOI: 10.1039/C4RA12111B |
| [56] | Xu Z, Liu W, Chen CC, et al. Enhanced α-zearalenol hydrolyzing activity of a mycoestrogen-detoxifying lactonase by structure-based engineering.ACS Catalysis, 2016, 6(11): 7657–7663.DOI: 10.1021/acscatal.6b01826 |
| [57] | Takahashi-Ando N, Ohsato S, Shibata T, et al. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea.Appl Environ Microbiol, 2004, 70(6): 3239–3245.DOI: 10.1128/AEM.70.6.3239-3245.2004 |
| [58] | Takahashi-Ando N, Tokai T, Hamamoto H, et al. Efficient decontamination of zearalenone, the mycotoxin of cereal pathogen, by transgenic yeasts through the expression of a synthetic lactonohydrolase gene.Appl Microbiol Biot, 2005, 67(6): 838–844.DOI: 10.1007/s00253-004-1816-y |
| [59] | Higa-Nishiyama A, Takahashi-Ando N, Shimizu T, et al. A model transgenic cereal plant with detoxification activity for the estrogenic mycotoxin zearalenone.Transgenic Res, 2005, 14(5): 713–717.DOI: 10.1007/s11248-005-6633-2 |
| [60] | Cheng BC, Shi WT, Luo J, et al. Cloning of zearalenone-degraded enzyme gene (ZEN-jjm) and its expression and activity analysis.Chin J Agric Biotechol, 2010, 18(2): 225–230.(in Chinese). 程波财, 史文婷, 罗洁, 等. 玉米赤霉烯酮降解酶基因(ZEN-jjm)的克隆, 表达及活性分析.农业生物技术学报, 2010, 18(2): 225-230. |
| [61] | Tan QL, Xu F, Li P, et al. Construction of an ZEN-jjm expressing vector and its expression in Pichia pastoris.Chin J Microecol, 2010, 22(12): 1061–1064.(in Chinese). 谭强来, 徐锋, 黎鹏, 等. 玉米赤霉烯酮降解酶毕赤酵母表达载体的构建及其表达.中国微生态学杂志, 2010, 22(12): 1061-1064. |
| [62] | Liu HY, Sun CP, Wu SL, et al. Expression of zearalenone degrading enzyme gene from Gliocladium Roseum in Pichia pastoris GS115.J Chin Cereals Oils Association, 2011, 26(5): 12–17.(in Chinese). 刘海燕, 孙长坡, 伍松陵, 等. 玉米赤霉烯酮毒素降解酶基因的克隆及在毕赤酵母中的高效表达.中国粮油学报, 2011, 26(5): 12-17. |
| [63] | Wang YC, Wang Y, Jiang JP, et al. Enzymatic properties of zearalenone degradation enzyme.Cereal & Feed Industry, 2015, 12(12): 64–68.(in Chinese). 王义春, 王龑, 江均平, 等. 玉米赤霉烯酮降解酶酶学性质研究.粮食与饲料工业, 2015, 12(12): 64-68. |
| [64] | Xiang L, Wang Q, Zhou Y, et al. High-level expression of a ZEN-detoxifying gene by codon optimization and biobrick in Pichia pastoris.Microbiol Res, 2016, 193: 48–56.DOI: 10.1016/j.micres.2016.09.004 |
| [65] | Takahashi-Ando N, Ohsato S, Shibata T, et al. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea.Appl Environ Microb, 2004, 70(6): 3239–3245.DOI: 10.1128/AEM.70.6.3239-3245.2004 |
| [66] | Yang WC, Hsu TC, Cheng KC, et al. Expression of the Clonostachys rosea lactonohydrolase gene by Lactobacillus reuteri to increase its zearalenone-removing ability.Microb Cell Fact, 2017, 16(1): 69–80.DOI: 10.1186/s12934-017-0687-8 |
| [67] | Altalhi AD, El-Deeb B. Localization of zearalenone detoxification gene(s) in pZEA-1 plasmid of Pseudomonas putida ZEA-1 and expressed in Escherichia coli.J Hazard Mater, 2009, 161(2): 1166–1172. |
| [68] | Tang Y, Xiao J, Chen Y, et al. Secretory expression and characterization of a novel peroxiredoxin for zearalenone detoxification in Saccharomyces cerevisiae.Microbiol Res, 2013, 168(1): 6–11.DOI: 10.1016/j.micres.2012.08.002 |
