

辽宁师范大学 生命科学学院 辽宁省植物生物技术重点实验室,辽宁 大连 116081
收稿日期:2016-06-14;接收日期: 2016-09-19; 网络出版日期:2016-09-26 基金项目:国家自然科学基金 (No. 31340052),辽宁省教育厅科技研究项目 (No. L201683655) 资助。
摘要: 研究已表明植物特有的一些NAC (NAM, ATAF1/2, CUC2) 转录因子可提高植物抗逆性,利用基因芯片技术筛选转SlNAC1基因拟南芥与野生型拟南芥间差异表达基因,能够为研究转基因拟南芥非生物胁迫抗性相关基因提供依据。结果显示,在转SlNAC1基因拟南芥43 604个基因中有3 046个差异表达2倍以上的基因。对差异表达5倍以上基因经过GO富集度统计学分析表明,细胞组分相关基因占33.05%;分子功能相关基因占33.95%;生物学过程相关基因占33.00%。对差异表达2倍以上基因进行KEGG信号通路分析,结果表明有2 431个基因涉及到88个不同的信号通路。通过筛选获得转基因拟南芥非生物胁迫抗性相关候选基因,为后续研究NAC转录因子的下游基因及其调控网络的构建提供方向和理论支撑。
关键词: 差异表达 转基因 非生物胁迫抗性 转录因子 GO分析 KEGG
Gene chip analysis of differentially expressed genes in transgenic SlNAC1 Arabidopsis
Yu Jingyang, Li Qianqian, Jiao Yang, Li Qiuli


College of Life Science, Liaoning Normal University, Dalian 116081, Liaoning, China
Received: June 14, 2016; Accepted: September 19, 2016; Published: 2016-September-26
Supported by:National Natural Science Fundation of China (No. 31340052), Scientific Research Project of Liaoning Province EducationDepartment (No. L201683655).
Corresponding authors:Qiuli Li. Tel/Fax: +86-411-85827073; E-mail: skyliqiuli@163.com
Abstract: Studies have shown that some plant-specific NAC (NAM, ATAF1/2, CUC2) transcription factors may increase plants resistance to stress. We screened the genes differentially expressed in transgenic SlNAC1 Arabidopsis compared to the wild type by cDNA microarry, to provide scientific basis for studying the genes related to abiotic stress responses in transgenic Arabidopsis. There were 3 046 genes differentially expressed more than twice in the total 43 604 genes of transgenic SlNAC1 Arabidopsis. Gene ontology analysis was used on genes differentially expressed more than five-fold. Genes relevant to cellular components occupied 33.05%, genes correlated with molecular function accounted for 33.95% and genes pertinent to biological process constituted a 33.00% portion. The genes differentially expressed more than twice were processed through kyoto encyclopedia of genes and genomes pathways enrichment (KEGG) analysis. The total 2 431 genes were involved in 88 different signaling pathways. The screened genes related to abiotic stress responses provide direction and theoretical support for the following research on the downstream genes regulated by NAC and construction of the regulatory networks.
Key words: differential expression transgenosis abiotic stress resistance transcription factor Gene ontology analysis KEGG
植物在生长过程中不可避免地会受到生物胁迫 (由生物引起,如病菌、虫害等) 或非生物胁迫 (由一系列过度或不足的物理、化学条件变化所引发的不利于植物生长发育的影响因素,如干旱、高温、冷害、冻害等),它们都能影响植物的生长、发育而降低产量。植物根据特定的环境胁迫进化出多种适应性的分子机制 (通过激活或抑制特定靶基因改变其表达量)[1]来响应不同的非生物胁迫。植物感知到胁迫后通过信号转导来激活胁迫响应相关基因的表达,参与胁迫应答基因表达调控的核心组件包括激酶、磷酸酶和转录因子 (Transcription factor) 等[2]。转录因子是能够结合在某基因上游特异核苷酸序列上的蛋白质,这些蛋白质能调控其下游基因的转录。WRKY、MYB、bZIP和NAC等多种转录因子都能参与调控植物生长发育并提高植物抗逆性。
Souer等最先在矮牵牛Petunia hybrid V. 中分离出NAC转录因子,命名为NAM[3]。1997年Aida等[4]报道了NAC (NAM,ATAF1/2和CUC) 转录因子的结构域,并命名为NAC。NAC转录因子是具有多种生物学功能的植物特有的转录因子[5],具有参与调控植物发育 (胚、茎尖、侧根形成,生长素信号转导和叶片衰老等)[6-8]、抵抗生物胁迫[9]与非生物胁迫 (干旱、高盐、低温、高温、高光、ABA)[10-12]等功能。在植物响应生物、非生物胁迫过程中,NAC转录因子通过调控多个下游基因的表达提高植物抗逆性。拟南芥Arabidopsis thaliana L. NAC家族中的ANAC019、ANAC055和ANAC072转录因子通过调控MYB2和MYB108转录因子的表达响应胁迫、调节叶片的衰老[13]。ANAC096转录因子通过调控RD29A的表达提高转基因拟南芥干旱和渗透胁迫抗性[2]。ChIP-Seq数据显示大豆 Glycine max cv. Williams 中有72个基因可能受NAC转录因子调控,RNA-Seq结果表明受NAC转录因子调控的差异表达基因包括脂肪氧化酶基因、果胶甲酯酶抑制剂基因、DEAD/DEAH解旋酶基因等[14]。
实验室前期从辽宁碱蓬Suaeda liaotungensis K. 中克隆了SlNAC1基因,通过农杆菌转化法获得了转SlNAC1基因拟南芥,转SlNAC1基因提高了拟南芥在盐、干旱、冷胁迫下的存活率[15],认为转SlNAC1基因拟南芥抗逆性增强。本文以转SlNAC1基因拟南芥为实验材料,通过基因芯片技术筛选其与野生型拟南芥的差异表达基因,希望从差异表达基因中确定NAC转录因子调控的下游基因,进而分析NAC转录因子的调控 网络。
1 材料与方法1.1 实验材料野生型拟南芥Arabidopsis thaliana L. 为哥伦比亚生态型 (Columbia-0) (命名为WT) 和转SlNAC1基因拟南芥 (命名为L2)。
1.2 拟南芥幼苗培养与取材 取适量的WT和L2种子分别用70%乙醇浸泡30 s,ddH2O冲洗3遍,再用10%次氯酸钠浸泡5 min,ddH2O冲洗7遍。灭菌后的种子分别播种于MS培养基中,置于培养箱中培养 (22 ℃,光照16 h,黑暗8 h),至6片叶子后将其转移至含有蛭石的培养钵中。用清水浇灌幼苗,每5 d浇灌1次,20 d后取样。分别从WT和L2拟南芥株系的3个植株中取材,每株取1片叶子,每个株系共取3片叶子,迅速置于液氮中,-80 ℃保存。
1.3 芯片实验 采用TaKaRa RNAiso Plus试剂盒分别提取WT和L2叶片总RNA。NanoDrop ND-2000分光光度计及Agilent Bioanalyzer 2100检测总RNA的质量,对总RNA中的mRNA进行放大、标记,用RNeasy Mini Kit纯化标记后的cRNA。按照Agilent表达谱芯片配套的杂交标准流程和配套试剂盒进行杂交和洗涤。完成杂交的芯片采用Agilent Microarray Scanner进行扫描。用Feature Extraction software 10.7读取数据,最后对质控合格的数据采用Gene Spring Software 12.6.1进行归一化处理,算法为Quantile。以Fold change≥2或Fold change≤0.5为标准筛选差异表达基因。本实验由上海伯豪生物技术有限公司完成,所用芯片为Agilent拟南芥全基因组4×44K芯片,每个样品各做一张芯片。在Agilent表达谱芯片实验中,用10次重复探针点信号的CV值来计算芯片的稳定性和技术的稳定性,其质控标准是平均CV<10%,相关系数r2>0.95。
1.4 聚类分析 对差异表达2倍以上基因中的WRKY、DREB和MYB等转录因子运用SAS在线分析系统进行聚类分析,并用Tree View软件来显示聚类结果。
1.5 GO富集分析对差异表达5倍以上基因进行GO富集分析,对细胞组分、分子功能和生物学过程中差异表达基因个数进行统计并绘图。满足P<0.05条件的GO term定义为在差异表达基因中显著富集的GO term。
1.6 KEGG富集分析对差异表达2倍以上的基因进行KEGG富集分析,把差异表达显著的基因通路进行富集(筛选的标准为P<0.05),统计涉及该通路的差异表达基因的个数、确定通路中的差异基因并对其进行初步分析。
1.7 实时荧光定量PCR 为验证基因芯片结果的可靠性,根据差异表达基因的功能及差异表达倍数,选择5个差异表达基因 (上调表达基因4个,下调表达基因1个)进行实时荧光定量PCR。根据差异表达基因序列设计并合成引物 (由上海生工生物有限公司完成)。使用PrimeScript? RT reagent Kit with gDNA Eraser (Perfect Real Time) 试剂盒(TaKaRa) 进行反转录。使用TaKaRa SYBR? Premix Ex TaqTM Ⅱ (Tli RNaseH Plus) 试剂盒和Thermal Cycler Dice Real time (TaKaRa) 进行实时荧光定量PCR反应,以Atactin-2基因 (NM_112764)作为内参基因。2-ΔΔCt法计算目的基因的相对表达量。每个样品重复3次。
2 结果与分析2.1 样本总RNA提取与质量检测 提取样本叶片总RNA并测定吸光度,WT样本叶片总RNA浓度100.6 ng/μL,体积15 μL,总量1.51 μg,A260/A280=1.85,28S/18S=1.4;L2样本叶片总RNA浓度63.6 ng/μL,体积15 μL,总量0.95 μg,A260/A280=1.88,28S/18S=1.6。样本叶片总RNA的A260/A280吸光度比值均在1.8至2.0之间;样本总RNA质检图 (图 1) 显示,各条带清晰完整,且28S和18S条带亮度接近2∶1,说明提取的总RNA纯度和完整性均较好,质量检测合格,达到后续的芯片实验要求。
 |
| 图 1 WT与L2样本总RNA Agilent Bioanalyzer 2100质检图 Figure 1 QC results of total RNA samples of WT and L2 Arabidopsis by Agilent Bioanalyzer 2100. Nt: nucleotide. FU: fluorescence value. (A) The fluorescence value of ladder under different nucleotide. (B) The fluorescence value of WT under different nucleotide. (C) The fluorescence value of L2 under different nucleotide. (D) QC results of total RNA samples of WT and L2. |
| 图选项 |
2.2 芯片质控与扫描 WT基因芯片CV值为7.35%,检出率为58.67%;L2基因芯片CV值为6.02%,检出率为59.01%。对芯片进行荧光扫描,扫描图 (图 2)中杂交信号清晰、均衡且背景清晰,所以芯片结果真实可靠。
 |
| 图 2 WT (左) 与L2 (右) 基因芯片扫描图 Figure 2 The results of gene chip scanning of WT (left) and L2 (right) Arabidopsis. |
| 图选项 |
在野生型和转SlNAC1基因拟南芥表达谱比较的线性归一化后的散点图 (图 3) 中,X轴为WT荧光强度值,Y轴为L2荧光强度值,每个数据点代表芯片上的一个基因点的杂交信号。绝大多数的点密集分布于y=x直线附近灰色区域中,表示在WT与L2之间没有明显的差异,信号值差异Fold Change=1。落在图形中位线两侧靠近Y轴的点表明该基因在L2中表达上调,靠近X轴的点表明该基因在L2中表达下调,信号值差异Fold Change>2。
 |
| 图 3 WT与L2芯片数据散点图 Figure 3 Scatter plot of cluster expression between WT and L2. |
| 图选项 |
2.3 差异表达基因统计 在转SlNAC1基因拟南芥43 604个基因中检测到3 046个基因差异表达2倍以上,上调与下调2倍以上基因的详细分布情况见表 1。其中,9个基因上调30倍以上 (表 2),4个基因下调300倍以上 (表 3)。
表 1 转SlNAC1基因拟南芥差异表达基因上调、下调倍数分布个数表Table 1 The number of genes up-regulated and down-regulated in transgenic SlNAC1 Arabidopsis from twice to the most compared to WT
| Fold change | Up-regulated gene number | Down-regulated gene number |
| 2<FC≤5 | 1 213 | 1 063 |
| 5<FC≤10 | 245 | 255 |
| 10<FC≤30 | 87 | 133 |
| 30<FC≤50 | 8 | 20 |
| 50<FC≤100 | 0 | 10 |
| 100<FC≤300 | 1 | 7 |
| 300<FC | 0 | 4 |
| Total | 1 554 | 1 492 |
| 3 046 |
表选项
表 2 转SlNAC1基因拟南芥差异表达上调30倍以上基因Table 2 The genes up-regulated more than 30 times in transgenic SlNAC1 Arabidopsis compared to WT
| Gene symbol | Probe name | Fold change | GenBank Accession No. | Description |
| AT2G14247 | A_84_P564098 | 130.82 | NM_201723 | Arabidopsis uncharacterized protein mRNA |
| AT2G30766 | A_84_P758588 | 44.70 | NM_001124947 | Arabidopsis uncharacterized protein mRNA |
| CXXS2 | A_84_P804145 | 41.96 | NM_129642 | Arabidopsis thioredoxin-like protein CXXS2 mRNA |
| AT3G16670 | A_84_P193394 | 38.62 | NM_112540 | Arabidopsis pollen Ole e 1 allergen and extensin family protein mRNA |
| ABI4 | A_84_P23876 | 36.68 | NM_129580 | Arabidopsis ethylene-responsive transcription factor ABI4 mRNA |
| AT1G47395 | A_84_P549811 | 35.45 | NM_179449 | Arabidopsis uncharacterized protein mRNA |
| SCRL24 | A_84_P723202 | 33.36 | NM_001036699 | Arabidopsis SCR-like 24 mRNA |
| AT4G28780 | A_84_P13857 | 32.33 | NM_119022 | Arabidopsis GDSL esterase/lipase mRNA |
| bZIP5 | A_84_P17486 | 31.72 | NM_114836 | Arabidopsis basic leucine-zipper 5 mRNA |
表选项
表 3 转SlNAC1基因拟南芥差异表达下调300倍以上基因Table 3 The genes down-regulated more than 300 times in transgenic SlNAC1 Arabidopsis compared to WT
| Gene symbol | Probe name | Fold change | GenBank Accession No. | Description |
| VSP1 | A_84_P97476 | 9.25E-4 | NM_122387 | Arabidopsis acid phosphatase VSP1 mRNA |
| VSP2 | A_84_P808818 | 2.21E-3 | NM_001036860 | Arabidopsis acid phosphatase VSP2 mRNA |
| AT4G11320 | A_84_P20420 | 2.49E-3 | NM_117203 | Arabidopsis putative cysteine proteinase mRNA |
| LTP3 | A_84_P20704 | 3.19E-3 | NM_125323 | Arabidopsis non-specific lipid-transfer protein 3 mRNA |
表选项
2.4 聚类分析 在差异表达2倍以上基因中,通过关键词“WRKY”、“DREB”和“MYB”等检索出61个转录因子 (22个WRKY、5个bZIP、1个DREB、4个ERF、21个MYB、2个MYC、6个RAP) 进行层级聚类分析。层级聚类分析图 (图 4) 中黑色代表该基因在两个样本中表达水平没有变化,左上和右下的深色区域代表升高,右上和左下的浅色区域代表降低;颜色的深浅程度代表基因表达水平升高或降低的程度,可明显看出深色、浅色形成显著的4大块,说明两组形成了明显的聚类群。层级聚类分析显示,基因表达存在着明显不同的聚类。
 |
| 图 4 61个差异表达转录因子聚类热图 Figure 4 The cluster heatmap of 61 differential expression transcription factors. |
| 图选项 |
2.5 GO富集分析对转SlNAC1基因拟南芥差异表达5倍以上的基因 (770个) 进行GO富集分析,按主要的功能分类,细胞组分 (GO: 0005575 cellular component) 相关基因586个,占33.05%;分子功能 (GO: 0003674 molecular function) 相关基因602个,占33.95%;生物学过程 (GO: 0008150 biological process) 相关基因585个,占33.00%。细胞组分主要涉及细胞、细胞成分和细胞器等;分子功能主要涉及参与催化活性、结合和转录调节活性等;生物学过程主要涉及细胞过程、新陈代谢和响应刺激等 (图 5)。细胞组分相关基因经富集统计差异不显著 (P>0.05)。分子功能相关基因中,238个差异表达基因具有催化活性相关功能 (GO: 0003824 catalytic activity),富集统计差异极显著 (P=0.0017);62个差异表达基因具有转录调节活性相关功能 (GO: 0030528 transcription regulator activity),富集统计差异显著 (P=0.0132);14个差异表达基因具有酶调节活性功能 (GO: 0030234 enzyme regulator activity),富集统计差异显著 (P=0.0342)。生物学过程相关基因中,142个差异表达基因响应刺激 (GO: 0050896 response to stimulus),富集统计差异极显著 (P=0);43个差异表达基因参与多个生物学过程 (GO: 0051704multi-organism process),富集统计差异极显著 (P=0)。
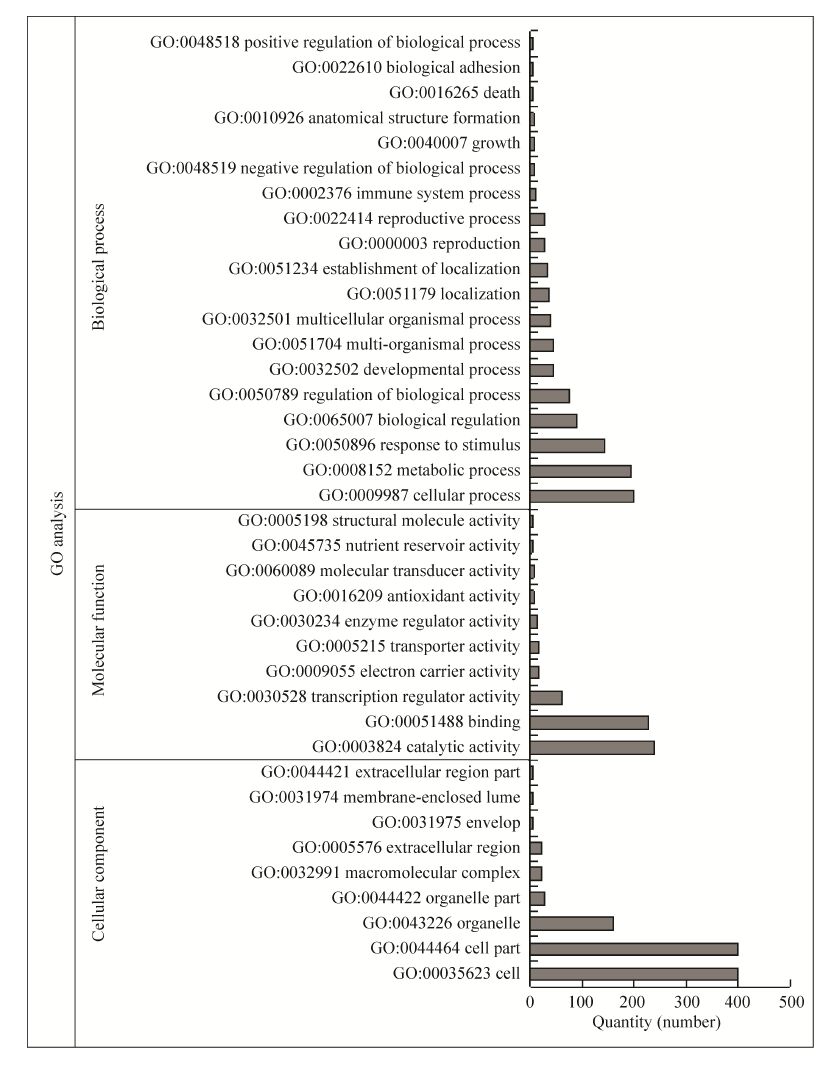 |
| 图 5 差异表达基因GO分析图 Figure 5 GO analysis graph of differentially expressed genes. |
| 图选项 |
2.6 KEGG信号通路分析对3 046个差异表达基因进行KEGG信号通路分析,这些基因涉及到88个富集但差异显著性不同的信号通路,其中差异极显著 (P<0.01)的信号通路有7个 (表 4),差异显著(0.01<P<0.05) 的信号通路有8个。
表 4 差异表达基因KEGG富集分析 (P<0.01)Table 4 KEGG analysis of differentially expressed genes (P<0.01)
| Pathway name | Total | Hits | P-value | Q-value | Representative differentially expressed genes |
| Plant hormone signal transduction | 232 | 40 | 0.0 | 3.0E-4 | ARR15,PYL6,PAN,IAA19,MYC2,TCH4 |
| Plant-pathogen interaction | 148 | 27 | 1.0E-4 | 0.0023 | CAM2,CPK24,MSS3,MYC2,TCH2,WRKY22 |
| Phenylalanine metabolism | 92 | 19 | 2.0E-4 | 0.0045 | 4CL3,AT2G34060,HPA1,PAL2,PRX33 |
| Phenylpropanoid biosynthesis | 109 | 19 | 0.0013 | 0.0178 | 4CL3,AT5G51890,HCT,PAL2,PRX33,PRXR1 |
| DNA replication | 45 | 11 | 0.0014 | 0.0178 | DPB2,ICU2,PCNA1,POLA2,POLD3,RPA2 |
| Lysine biosynthesis | 19 | 6 | 0.0061 | 0.0656 | AT3G57560,ALDH7B4,AT2G44040,AT3G14390 |
| Stilbenoid,diarylheptanoid and gingerol biosynthesis | 67 | 12 | 0.0078 | 0.0721 | HCT,PAD3,CYP91A2,BT2,CYP71B12,CYP81F2 |
表选项
2.7 实时荧光定量PCR验证差异表达基因为了验证基因芯片结果的可靠性,本研究挑选了5个差异表达基因 (上调表达基因4个,下调表达基因1个) 进行实时荧光定量PCR验证,结果显示,虽然各基因的表达倍数在基因芯片和实时荧光定量PCR中不同,但是总体趋势相同 (表 5),因此基因芯片的结果可靠。
表 5 实时荧光定量PCR与基因芯片的基因表达比较Table 5 The comparison of gene expression level between qRT-PCR and gene chip
| Gene symbol | GenBank Accession No. | Sequence of primers (5′-3′) | Product length (bp) | Fold change gene chip | Fold change qRT-PCR |
| Actin2 | NM_112764 | F: CAACCGGTTAGTACATTTTAGGC R: GTAAGGTCACGTCCAGCAAG | 193 | ||
| WRKY22 | NM_116355 | F: AAGCCACAGAACCAGAAACG R: TTGGGTGAAGAAACGAACCT | 116 | 6.80 | 8.39±0.55 |
| AT5G51890 | NM_124568 | F: ATTTCACGAGCCAACGAGAC R: CCACCTGAGAGCGTAACCAT | 119 | 4.82 | 4.15±0.56 |
| bHLH100 | NM_129689 | F: CACCGACCAAAACAGTAAGTCA R: TCAAGACATTCCCAAACGAA | 140 | 19.94 | 11.77±0.43 |
| MYB96 | NM_125641 | F: CCGCAACGATTAGCTTTTGT R: GGCCCTTTCTTCACTCCAAT | 108 | 2.17 | 4.32±0.60 |
| VSP1 | NM_122387 | F: ATCCGTTCCAGGGCTCAT R: GCAGTTGGGGTAGTTGATGG | 103 | 9.25E-4 | 2.91E-2±0.01 |
表选项
3 讨论 在植物生长发育和响应生物及非生物胁迫过程中,转录因子起着重要作用,通过调控多个下游基因的表达提高植物抗逆性。转SlNAC1基因拟南芥3 046个差异表达2倍以上基因中共筛选到61个转录因子。植物中的MYB转录因子参与植物体内的众多生理反应,能调控植物的生长发育,诱导植物参与非生物胁迫的应答。NAC转录因子ANAC019和ANAC055通过调控MYB2、MYB21、MYB108、MYB112和MYB116的表达提高拟南芥抗逆性[13],推测SlNAC1可能通过调控MYB6 (上调2.37倍)、MYB30 (上调3.23倍)、MYB96 (上调2.17倍)和MYB105 (上调4.57倍) 等MYB转录因子基因的表达提高转基因拟南芥抗逆性。WRKY是植物特有的转录因子之一,具有参与植物生长发育、生物和非生物胁迫响应和激素信号转导等多种生物学功能,拟南芥WRKY57转录因子参与调控外源植物激素茉莉酸和生长素所介导的植物叶片衰老信号途径之间的交叉调控通路[16],在转SlNAC1基因拟南芥中,SlNAC1调控WRKY22 (上调6.80倍)、WRKY28 (上调6.04倍)、WRKY53 (上调6.02倍) 和WRKY56 (上调6.04倍) 等的表达,共同调控拟南芥生长的抗逆性。
碱性亮氨酸拉链 (Basic region leucine zipper motif,bZIP) 类转录因子普遍存在于动植物及微生物中,参与植物生长、种子成熟、衰老等生物学过程,能提高植物抵抗各种不良环境 (病原体入侵、高温、冷害、高盐等) 的能力[17-18],推测SlNAC1通过调控bZIP5 (上调31.72倍) 等bZIP类转录因子基因的表达提高转基因拟南芥抗逆性。植物营养贮存蛋白质 (Vegetative storage proteins,VSP) 是植物防御相关的重要蛋白之一,在拟南芥中发现VSP1和VSP2两种,受机械创伤、茉莉酸、昆虫的咬食和渗透胁迫等诱导[19-20]。在转SlNAC1基因拟南芥中,VSP1和VSP2基因分别下调表达1081.32、452.33倍,有可能会导致转基因拟南芥防御能力降低。
KEGG富集分析表明,植物激素信号转导、植物病原体相互作用、苯丙氨酸代谢、DNA复制等7个信号通路富集度极显著。植物激素具有参与调控种子萌发、细胞分裂、组织和器官建成、开花与结实等作用[21],不同种类植物激素间相互作用对植物的生长发育至关重要。MYC2是bHLH (Basic-helix-loop-helix) 转录因子家族成员,参与调控茉莉酸信号通路与其他植物激素 (脱落酸、水杨酸、赤霉素和生长素)间的交叉对话,MYC2还调节茉莉酸介导的虫害和病原体防御反应[22]。MYC2参与植物激素信号转导通路,SlNAC1可能通过调控MYC2 (上调2.26倍) 的表达影响植物激素信号转导通路,影响转基因拟南芥的抗逆性。MYC2参与植物病原体相互作用通路,SlNAC1可能通过调控MYC2的表达影响植物病原体相互作用通路,增强转基因拟南芥的抗病性。SlNAC1与ANAC062和ANAC091同属于TIP亚家族[23],ANAC062、ANAC091能提高拟南芥抗病性[24-26],推测SlNAC1可能具有抗病功能。
目前关于NAC调控网络的研究报道较少,本文以转SlNAC1基因和野生型拟南芥为实验材料,利用基因芯片技术筛选出与胁迫相关的差异表达基因、转录因子及相关的代谢通路。SlNAC1通过调控这些相关基因的表达提高转基因拟南芥的抗逆性。
参考文献
| [1] | Wang ZY, Dane F. NAC (NAM/ATAF/CUC) transcription factors in different stresses and their signaling pathway.Acta Physiol Plant, 2013, 35(5): 1397–1408.DOI: 10.1007/s11738-012-1195-4 |
| [2] | Xu ZY, Kim SY, Hyeon DY, et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses.Plant Cell, 2013, 25(11): 4708–4724.DOI: 10.1105/tpc.113.119099 |
| [3] | Souer E, van Houwelingen A, Kloos D, et al. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries.Cell, 1996, 85(2): 159–170.DOI: 10.1016/S0092-8674(00)81093-4 |
| [4] | Aida M, Ishida T, Fukaki H, et al. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant.Plant Cell, 1997, 9(6): 841–857.DOI: 10.1105/tpc.9.6.841 |
| [5] | Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes.Science, 2000, 290(5499): 2105–2110.DOI: 10.1126/science.290.5499.2105 |
| [6] | Takasaki H, Maruyama K, Takahashi F, et al. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence.Plant J, 2015, 84(6): 1114–1123.DOI: 10.1111/tpj.13067 |
| [7] | He X, Qu BY, Li WJ, et al. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield.Plant Physiol, 2015, 169(3): 1991–2005. |
| [8] | Ning YQ, Ma ZY, Huang HW, et al. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14.Nucleic Acids Res, 2015, 43(3): 1469–1484.DOI: 10.1093/nar/gku1382 |
| [9] | Wang FT, Lin RM, Feng J, et al. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana.Front Plant Sci, 2015, 6: 108–124. |
| [10] | Sakuraba Y, Kim YS, Han SH, et al. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP.Plant Cell, 2015, 27(6): 1771–1787.DOI: 10.1105/tpc.15.00222 |
| [11] | Sakuraba Y, Piao W, Lim JH, et al. Rice ONAC106 inhibits leaf senescence and increasessalttolerance and tiller angle.Plant Cell Physiol, 2015, 56(12): 2325–2339.DOI: 10.1093/pcp/pcv144 |
| [12] | Yang X, Hu YX, Li XL, et al. Molecular characterization and function analysis of SlNAC2 in Suaeda liaotungensis K.Gene, 2014, 543(2): 190–197.DOI: 10.1016/j.gene.2014.04.025 |
| [13] | Hickman R, Hill C, Penfold CA, et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves.Plant J, 2013, 75(1): 26–39.DOI: 10.1111/tpj.2013.75.issue-1 |
| [14] | Shamimuzzaman M, Vodkin L. Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq.BMC Genomics, 2013, 14(1): 477–493.DOI: 10.1186/1471-2164-14-477 |
| [15] | Li XL, Yang X, Hu YX, et al. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance.Plant Cell Rep, 2014, 33(5): 767–778.DOI: 10.1007/s00299-014-1602-y |
| [16] | Jiang YJ, Liang G, Yang SZ, et al. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence.Plant Cell, 2014, 26(1): 230–245.DOI: 10.1105/tpc.113.117838 |
| [17] | Nijhawan A, Jain M, Tyagi AK, et al. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice.Plant Physiol, 2008, 146(2): 333–350. |
| [18] | Jakoby M, Weisshaar B, Drge-Laser W, et al. bZIP transcription factors in Arabidopsis.Trends Plant Sci, 2002, 7(3): 106–111.DOI: 10.1016/S1360-1385(01)02223-3 |
| [19] | Berqer S, Mitchell-Olds T, Stotz HU. Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana.Physiol Plant, 2002, 114(1): 85–91.DOI: 10.1046/j.0031-9317.2001.1140112.x |
| [20] | Liu Y, Ahn JE, Datta S, et al. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase.Plant Physiol, 2005, 139(3): 1545–1556.DOI: 10.1104/pp.105.066837 |
| [21] | Waadt R, Hsu PK, Schroeder JI. Abscisic acid and other plant hormones: methods to visualize distribution and signaling.Bioessays, 2015, 37(12): 1338–1349.DOI: 10.1002/bies.v37.12 |
| [22] | Kazan K, Manners JM. MYC2: the master in action.Mol Plant, 2013, 6(3): 686–703.DOI: 10.1093/mp/sss128 |
| [23] | Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana.DNA Res, 2003, 10(6): 239–247.DOI: 10.1093/dnares/10.6.239 |
| [24] | Truman W, de Zabala MT, Grant M. Type III effectors orchestrate a complex interplay between transcription networks to modify basal defence responses during pathogenesis and resistance.Plant J, 2006, 46(1): 14–33.DOI: 10.1111/tpj.2006.46.issue-1 |
| [25] | Yang ZT, Lu SJ, Wang MJ, et al. A plasma membrane-tethered transcription factor, NAC062/ ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis.Plant J, 2014, 79(6): 1033–1043.DOI: 10.1111/tpj.12604 |
| [26] | Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus.Plant Cell, 2000, 12(10): 1917–1926.DOI: 10.1105/tpc.12.10.1917 |
