杨梓琪1,2, 程晓钰1,2, 王红梅1,2

 , 曾智霖1,2, 刘晓燕1,2, 曹静1,2, 苏春田3, 黄奇波3
, 曾智霖1,2, 刘晓燕1,2, 曹静1,2, 苏春田3, 黄奇波3 1. 中国地质大学(武汉)环境学院, 湖北 武汉 430074;
2. 中国地质大学(武汉), 生物地质与环境地质国家重点实验室, 湖北 武汉 430074;
3. 中国地质调查局岩溶地质研究所, 广西 桂林 541000
收稿日期:2021-06-06;修回日期:2021-09-07;网络出版日期:2021-09-28
基金项目:国家自然科学基金(91951208);广西自然科学基金(2020GXNSFAA297025);中国地质调查项目(DD20190343)
*通信作者:王红梅, E-mail: hmwang@cug.edu.cn; wanghmei04@163.com.
摘要:[目的] 洞穴被认为是黑暗、寡营养的极端环境,是研究深地生物圈的天然实验室。洞穴内部小生境丰富,不同洞穴水文条件和环境因子等差异大,尽管微生物群落在不同的洞穴中均显示出较强的生境特异性,但对不同相态(固相和液相)环境样本中微生物群落的环境驱动机制以及群落构建的生态学过程的认识却十分薄弱。为了回答上述科学问题。[方法] 本文选择了桂林地区典型的喀斯特洞穴罗汉肚洞,针对洞穴中不同生境(岩壁、沉积物、水潭积水、滴水和地下河河水)进行系统采样以及16S rRNA扩增子的高通量测序分析和理化参数的测试。[结果] 结果表明洞穴中不同生境微生物群落结构具有显著的生境特异性。岩壁样品以放线菌门(Actinobacteria)为优势类群,沉积物中的优势类群则为酸杆菌门(Acidobacteria),所有水样微生物群落均以g-变形菌纲(Gammaproteobacteria)为主。温度、风化指数以及SO42-浓度显著影响罗汉肚岩壁和沉积物等固相样本中微生物的群落结构,其中USCg和假单胞菌属(Pseudomonas)与温度呈正相关,假诺卡氏菌属(Pseudonocardia)、土壤红色杆形菌(Solirubrobacter)和芽单胞菌属(Gemmatimonas)则与温度显著负相关。而滴水、水潭积水以及地下河河水等液相样本中微生物群落则与电导率(EC)和溶解氧(DO)的含量显著相关。细菌群落的共生网络具有明显的模块性,不同微生物类群间以正相关的合作关系为主,以共同抵抗洞穴中的极端条件。固相样本中群落构建确定性过程(48.75%)与随机性过程(51.25%)的贡献基本相等,但液相样本中微生物的群落构建则以随机过程占主导(64.76%)。[结论] 本研究结果首次揭示了洞穴微生物在不同相态样品中微生物群落的分布规律以及环境驱动机制、网络互作方式以及群落构建等生态学过程的差异,为深刻认识洞穴这一深地生物圈的微生物空间分布特征及微生物与环境之间以及不同微生物类群之间的相互作用提供了新的视角。
关键词:极端环境喀斯特洞穴细菌群落环境驱动机制共生网络群落构建
Environmental driving mechanisms and community assembly process of bacterial communities in the Luohandu Cave, Guilin, Guangxi Province, China
Ziqi Yang1,2, Xiaoyu Cheng1,2, Hongmei Wang1,2

 , Zhilin Zeng1,2, Xiaoyan Liu1,2, Jing Cao1,2, Chuntian Su3, Qibo Huang3
, Zhilin Zeng1,2, Xiaoyan Liu1,2, Jing Cao1,2, Chuntian Su3, Qibo Huang3 1. School of Environmental Studies, China University of Geosciences, Wuhan 430074, Hubei Province, China;
2. State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan 430074, Hubei Province, China;
3. Institute of Karst Geology of China Geological Survey, Guilin 541000, Guangxi Zhuang Autonomous Resion, China
Received: 6 June 2021; Revised: 7 September 2021; Published online: 28 September 2021
*Corresponding author: Hongmei Wang, E-mail: hmwang@cug.edu.cn; wanghmei04@163.com.
Foundation item: Supported by the National Natural Science Foundation of China (91951208), by the Guangxi Natural Science Foundation (2020GXNSFAA297025) and by the Geological Survey Project of China (DD20190343)
Abstract: [?] Caves are extreme environments with permanent darkness and limited nutrients, which serve as natural laboratories to study the subsurface deep biosphere. Although microbial communities have demonstrated strong niche specificity in different caves, the environmental driving mechanism of microbial communities and the ecological processes responsible for community assembly in different niches particularly those in different substrates e. g. solid versus liquid samples were poorly understood. [Objective] Here we aim to explore the environmental driving mechanisms and community assembly process of bacterial communities in different substrates e. g. solid versus liquid samples combined with physicochemical properties. [Methods] To this end, a karst cave, the Luohandu Cave in Guilin city, Guangxi province, locating in the typical karst region in southwestern China, was selected for a systematic investigation of microbial communities. Solid samples (weathered rocks and sediments) and liquid samples (dripping water, pool water and ground river water samples) were collected along the cave and subjected to high-throughput sequencing of 16S rRNA gene. Meanwhile, physicochemical properties of these samples were analyzed. [Results] Results showed strong niche specificity of microbial communities in the Luohandu cave. Actinobacteria and Acidobacteria dominated bacterial communities of weathered rocks and sediments, respectively. In contrast, water samples were dominated by γ-Proteobacteria. Redundancy analysis (RDA) between microbial communities and environmental variables demonstrated that temperature, weathering index and SO42- concentration significantly affect microbial communities in solid samples. USCγ and Pseudomonas positively associated with temperature, while Pseudonocardia, Solirubrobacter and Gemmatimonas negatively correlated with temperature. However, microbial communities in water samples were significantly controlled by electrical conductivity (EC) and dissolved oxygen (DO) as indicated by RDA. The co-occurrence network of bacterial communities was characterized by a good modularity with nodes from liquid samples locating in the same module, indicating a niche preference. Positive links dominated in the network, suggesting a corporative strategy among different microbial groups to survive in caves. Different ecological processes were found to be responsible for bacterial community assembly in different substrates. Deterministic process (48.75%) and stochastic process (51.25%) contributed almost equally to bacterial community assembly in solid samples, whereas stochastic process (64.76%) dominated the microbial community assembly in liquid samples. As for the individual ecological processes, homogenizing dispersal, undominated and dispersal limit in stochastic process contributed 40.42%, 10.46% and 3.13% to community assembly respectively; whereas homogenous selection and heterogeneous selection within deterministic process contributed 26.75% and 21.99%, respectively, in solid samples. In liquid samples, individual processes contributed to community assembly in order from high to low were undominated (28.57%), homogenous selection (25.71%), homogenizing dispersal (24.76%), dispersal limitation (10.48%), and heterogeneous selection (9.52%). [Conclusion] Our results for the first time revealed the different environmental driving mechanisms and different community assembly processes underlining microbial communities living in different substrates (solid versus liquid). This study offers a new window to understand the interactions among microbes and their environments, and interactions among different microbial groups in the subsurface biosphere in caves.
Keywords: extreme environmentkarst cavesbacterial communitiesenvironmental driving mechanismco-occurrence networkcommunity assembly
喀斯特地貌是由可溶性岩石在地下水和地表水的溶蚀与沉淀,侵蚀与沉积,以及重力崩塌、坍塌、堆积等作用形成的地貌。喀斯特地貌分布广泛,约占地球无冰区面积的20%[1-2],中国西南喀斯特地区(97°39′E-117°18′E,23°41′N-29°15′N)是世界上最大的喀斯特地貌分布区之一[3]。洞穴是一种典型的喀斯特地貌,地理位置相对隔离,长期黑暗、缺乏直接来自光合作用的有机质、湿度大(接近饱和)、温度变幅小、营养匮乏,被认为是一种极端的地下环境。洞穴与外部环境通过洞穴入口或岩溶地下水系统相连接,洞穴内外源物质(沉积物、有机颗粒等)的输入主要通过上覆岩层的裂隙水、地下河以及空气等携带进入洞穴[4],这些外源有机物的输入为异养微生物的生长提供了碳源。虽然洞穴中总有机碳含量极低,一般低于2 mg/L,但却孕育着大量的微生物,洞穴每克岩石中微生物的平均数量可达106个细胞[5]。
微生物在洞穴的形成以及洞穴内元素的循环中均起着十分重要的作用。有研究证明,微生物群落与各种环境条件共同作用塑造了喀斯特地貌,促进了无机矿物的形成,加速碳酸钙的沉积[6-8]。通过对洞穴微生物群落在生物地球化学循环中的功能预测,发现微生物可能参与了洞穴中的碳、氮循环,并且参与氨基酸代谢与碳水化合物代谢的基因具有高表达性[9]。基于氨氧化功能基因的研究证实奇古菌与氨氧化细菌均在洞穴氨氧化中起着重要的作用[10]。近年来,地球化学证据表明,由于微生物对甲烷的氧化作用,地下岩溶洞穴还可能是大气甲烷潜在的汇[11],对全球的甲烷循环起着至关重要的作用。分子生物学的证据也显示高亲和力的大气甲烷氧化菌尤其是Upland Soil Cluster γ (USCγ)类群在洞穴岩壁及沉积物的甲烷氧化菌群落中占主导地位[12-13]。最近研究者从贵州的两个喀斯特洞穴的岩石、沉积物和水样中分离培养出3562株细菌,隶属于329个种(包含14个新种)、102个属。结合数据库中已有的洞穴微生物的单株基因组以及对14个新种的基因组测序,发现了新的固氮微生物Azospirillum sp.和烷烃氧化微生物Oleomonas sp.,为认识洞穴微生物参与洞穴的碳、氮循环提供了新的证据[14]。
近年来人们对微生物在洞穴不同生境中的分布特征及其环境驱动机制的研究上取得了长足的进展。对洞穴岩壁、沉积物、滴水、岩石表面生物席、石笋表面生物膜、空气等多种生境微生物群落的研究表明,细菌群落具有显著的生境特异性,且pH和TOC是控制群落空间分布的重要因子[15-16]。同时,由于全球变暖效应以及微生物对温度的敏感性,温度对洞穴微生物的影响也备受关注[17]。滴水作为连接洞穴外部与内部的重要“桥梁”,多年的监测结果表明滴水中的细菌群落结构的季节性变化主要受控于洞外温度[18]。此外有研究发现,微生物群落特征也可能受到地质作用的长期影响,并把Ti/Fe、Mg/Al以及Ca/Al等化学风化指标纳入到微生物与环境相互作用的评估范围[19],并证实喀斯特洞穴上覆土壤样本中细菌的群落组成显著受风化作用(Ca/Al、Mg/Al和Si)的影响[20]。以上结果为深刻认识洞穴系统中微生物群落的分布特征及环境驱动机制提供了重要依据。然而,洞穴内样本本身的性质对微生物群落的影响,尤其是样本的相态(如固相样本和液相样本)对微生物群落特征以及微生物群落构建的生态学过程的影响,目前还不得而知。已有研究表明,不同相态样本中的微生物群落受控于不同的环境因子。如pH是影响土壤微生物群落的重要因子[21],而盐度则是控制湖泊微生物群落的重要环境因子[22-23]。海水中微生物群落与盐度、降雨量、温度以及水质等环境因子密切相关[24],而海洋沉积物中微生物群落则受控于沉积物深度、pH和CO2浓度[25],同时沉积物年龄和孔隙水中作为电子受体的硫酸盐的有效性显著决定了微生物群落的结构[26]。但也有研究表明,泥炭沉积物和孔隙水中的微生物群落均受控于水位的变化[27]。因此寻找上述科学问题在洞穴这一特殊生态系统的答案将为深刻揭示洞穴深地生物圈的微生物群落特征及微生物群落形成的生态学过程提供重要的科学依据。
为此,本文选择桂林喀斯特洞穴罗汉肚洞为研究区,该洞地下水系既发育有经上覆岩层裂隙进入洞穴的滴水、洞穴内部的水潭积水以及流经洞内的地下河河水等不同类型的水体样品,也发育有沉积物、风化岩壁等固相样本,为研究不同相态样本的微生物群落提供了良好的载体。在该洞中系统采集不同小生境的样本并进行16S rRNA扩增子的高通量测序,进而阐明罗汉肚洞穴中不同生境、不同相态样本中微生物群落特征及其与环境因子之间的相互关系、不同微生物类群之间的相互作用,最终揭示固相样本和液相样本中微生物群落的环境驱动机制和群落构建的生态学过程。研究结果将为深刻认识洞穴深地生物圈的微生物生态学过程提供重要参考。
1 材料和方法 1.1 样品采样 罗汉肚洞(25°0′55.8″N,110°54′14.2″E)位于桂林市恭城北15 km的嘉会乡白燕村东部的岩溶峰丛山地与峰林平原的边缘,洞口位于峰丛边缘峰体的山脚,洞口标高232 m。洞穴发育在中泥盆统东岗岭组厚层状、生物碎屑白云岩、灰质白云岩中,洞长356 m,洞高2-12 m,洞宽4-25 m,上覆岩层厚度约3-136 m。洞穴内部常年黑暗,地理位置相对隔离,洞内受人类活动影响较小,洞穴内发育有地下河、滴水以及水潭积水等。
于2019年1月按25 m的间距对罗汉肚洞从洞口到洞内最深处不同生境的样品进行了系统采样(图 1),包括10个点位的岩壁样本,9个点位的沉积物样本(其中3号点位无沉积物)。4号点和7号点(图 1-D)接取滴水样品,于6号和8号点采集水潭积水(图 1-B,C)以及地下河河水样本。每个采样点的每种样本均设3个生物学重复。在本研究中,岩壁、沉积物、滴水、水潭积水和地下河河水生境分别标记为W、S、DW、PW和R。水样经0.22 μm的滤膜抽滤,将微生物细胞收集到滤膜上,保存于无菌的50 mL离心管中,所得无菌滤液酸化后用于后续阴阳离子的测定。所有固相样本及滤膜用干冰保存运回实验室,置于-80 ℃冰箱保存,用于后续DNA的提取;无菌滤液置于4 ℃冰箱保存,用于理化参数的测定。洞穴气体样本利用微型采样泵(HDP12-WZ05,大连海德科技有限公司,中国)采集至1 L气体采样袋中(MBT41,大连海德科技有限公司,中国),采样期间需保证无人为扰动。
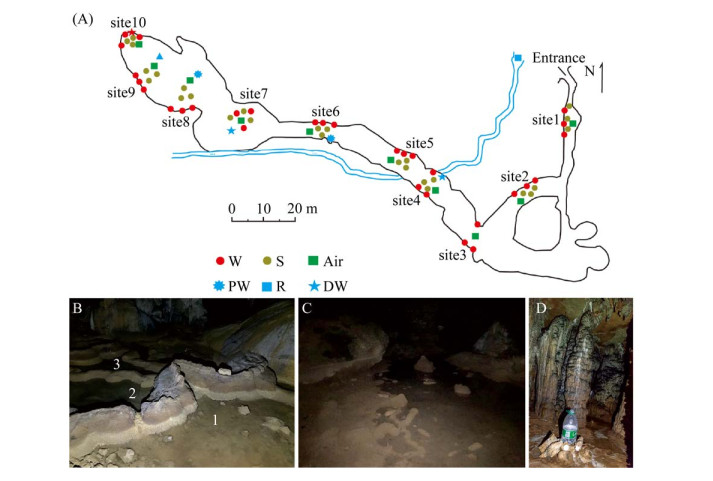 |
| 图 1 广西桂林罗汉肚洞穴采样点分布图 Figure 1 The sampling sites (A) and images of sampling site 6 (B), 8 (C) for pool water and 7 (D) for dripping water, respectively, in the Luohandu Cave, Guilin city, Guangxi province. |
| 图选项 |
1.2 物理化学参数测定 水样的温度、电导率、pH以及氧化还原电位用哈希水质测定仪(HACH,Loveland,CO,USA)在采样时进行原位测定,其他固相样本的理化参数如阴阳离子和总有机碳则在实验室对样品进行前处理,之后用相应的仪器进行测定。固相样品冷冻干燥后充分研磨,过200目筛。取6 g上述样本,加入到30 mL超纯水中振荡混匀10 min,以6800×g高速离心10 min。将上清液用0.22 μm滤膜过滤后用于检测阴阳离子。SO42-和Cl-等阴离子浓度用阴离子色谱仪(ICS-600,Thermo,USA)测定,Ca、Mg、Al、Fe等金属阳离子浓度则是由ICP-OES (iCAP 7600+,Thermo,USA)测定。固相样本总有机碳(TOC)用元素分析仪(Vario MACRO cube,Elementar,Germany)进行测定。
1.3 DNA提取及数据分析 利用Power Soil? DNA Isolation Kit (MOBIO,USA)提取0.5 g固相环境样品中的总DNA,同时利用Power Water? DNA Isolation Kit (MOBIO,USA)提取4 L水样中的总DNA,所得DNA浓度及质量由Nanodrop (ND2000,Thermofisher,USA)进行测定,高通量测序由上海派森诺生物科技有限公司在Illumina Miseq测序完成,测序引物为338F (5′-ACTCCTACGGGAGGCAGCA-3′)[28]/806R (5′-GGACTACHVGGGTWTCTAAT-3′)[29],测序的区间为细菌的16S rRNA V3-V4区。
获得的16S rRNA扩增子下机原始数据使用QIIME2平台分析,根据Q30剔除低质量序列及barcode后,使用DADA2去除嵌合序列,并划分ASVs (Amplicon Sequence Variants),生成Feature表格用于后续的数据分析[30]。使用Silva数据库(Release 132,http://www.arb-silva.de)和已发表数据库[31]对ASVs进行物种注释。并将所有样本序列进行均一化,每个样本中reads数均为29111,以避免因测序深度的不同而导致的群落多样性差异。在此条件下,使用R软件中的“vegan”包对细菌群落的Chao1、Shannon、Simpson、ACE等物种多样性指数进行计算。而基于Bray-Curtis距离矩阵对微生物群落进行非度量多维尺度分析(non-metric multidimensional scaling,NMDS)。使用Canoco5.0对微生物群落与环境因子进行冗余分析(redundancy analysis,RDA)。使用R中的“psych”包计算物种与环境因子的Spearman相关性。多反应置换法(multi response permutation procedure,MRPP)的计算结果用于评估不同生境间微生物群落组成差异的显著性。为减少复杂性,只保留相对丰度大于0.04%以上的ASV。使用R中的“psych”包计算得出ASV间相关系数矩阵,计算ASV间的Spearman相关系数,过滤相关系数r大于|0.6|、P值小于0.05为有效关系,基于以上参数生成共现网络(co-occurrence network)。导入Gephi (https://gephi.org)利用Fruchterman- Reingold布局进行可视化,并将其按模块和门进行着色。计算网络的网络级(平均节点度、聚类系数、平均路径长度、模块性、密度、直径、介数集中化和度集中化)和节点级(度、传递性、介数中心性和接近中心性)等拓扑特征参数。在Galaxy平台(http://huttenhower.sph.harvard.edu/galaxy)进行LEfSe分析。为了研究洞穴微生物群落的组装过程,用R的“picante”包计算最近分类单元指数(NTI)、最近分类单元距离(MNTD)、加权β平均最近分类单元距离(βMNTD)和加权β最近分类单元指数(βNTI)来衡量样品间的系统进化周转率[32-33]。当NTI值为正且平均值大于0 (P < 0.05)时,共现物种受系统发育的影响比过度扩散的影响更大[34]。如果|βNTI|值大于2,则确定过程是群落组成的关键组装过程。如果|βNTI|值低于2,则表明随机过程在细菌群落的形成中起着关键作用。βNTI值大于2或小于-2表示异质选择和同质选择是群落组装的主要过程。为了进一步辨别随机过程,基于群落的Bray-Curtis矩阵,通过“vegan”包计算了Raup-Crick矩阵(RCbray)。在1到+1范围内的RCbray值显示了零模型分布与观测到的Bray-Curtis之间的偏差。RCbray大于0.95,|RCbray|小于0.95以及RCbray小于-0.95分别表示扩散限制、不明确过程和均质扩散[32-33]。
2 结果 2.1 洞穴样品的理化性质 岩壁和沉积物样本中Ca/Mg具有显著差异,而K+、Cl-、SO42-以及Mg2+在各个生境间无显著差异。固相样品和水样的pH无显著差异,均呈弱碱性,水样中的pH整体比固体样本中低。电导率在不同来源的水样之间具有显著性差异,地下河水样电导率最低(139.67±13.65 μs/cm),水潭积水中电导率最高(585.83±18.42 μs/cm) (表 1)。而洞穴岩壁样品中的TOC含量(3.86±2.24%)平均比沉积物样品中的TOC含量(1.22±1.05%)高(表 1)。
表 1. 桂林罗汉肚洞穴样品的物理化学参数 Table 1. Physicochemical parameters of samples in the Luohandu Cave, Guilin City
| Niches | pH | TOC/% | K/(mg/L) | Cl-/(mg/L) | SO42-/ (mg/L) | Ca/Mg | EC/(μs/cm) | DO/(mg/L) | ORP/mV | T/℃ |
| W | 8.72±0.23 | 3.86±2.24 | 1.26±2.34 | 1.13±2.03 | 7.35±13.30 | 68.62±27.02a | - | - | - | - |
| S | 8.77±0.68 | 1.22±1.05 | 0.45±0.35 | 0.24±0.12 | 3.22±4.03 | 4.22±17.41b | - | - | - | - |
| PW | 7.66±0.18 | - | 0.10±0.03 | 1.15±0.05 | 6.70±1.10 | 1.50±0.95ab | 585.83±18.42a | 10.39±0.53 | 177.16±13.46 | 12.82±2.44 |
| DW | 8.29±0.10 | - | 0.58±1.16 | 0.85±0.22 | 7.52±1.13 | 10.71±4.18ab | 318.17±29.80b | 10.60±0.55 | 162.58±86.33 | 12.63±2.08 |
| R | 8.18±0.07 | - | 0.10±0.02 | 0.74±0.63 | 5.45±4.12 | 14.32±0.63ab | 139.67±13.65c | 9.93±0.08 | - | 15.30±0.50 |
| Statistical significance is assessed by one-way ANOVA, P < 0.05. Different lowercase letters shows statistically significant difference. | ||||||||||
表选项
2.2 洞穴不同生境微生物群落的结构特征以及指示类群 罗汉肚洞穴微生物群落的Alpha多样性随生境而变化(表 2)。岩壁和沉积物的Shannon指数高(8.43±0.74和9.29±0.62),而液相样本样品的Shannon指数相对较低[(6.95±2.18)-(8.18±0.09)] (表 2)。而滴水和地下河河水则具有较高的Chao1指数(2807±882) (表 2),而水潭积水中微生物的ACE、Chao1以及Shannon指数等均显著低于滴水和地下河水(表 2)。除水潭积水样本外,其他液相样本中微生物群落相较其他生境物种丰富度更高,但物种多样性则较低。
表 2. 桂林罗汉肚洞不同生境样品的α-多样性指数 Table 2. The α-diversity indices of samples among different niches in the Luohandu Cave, Guilin City
| Niches | Community richness and diversity index | |||
| ACE | Chao1 | Shannon | Simpson | |
| W | 1448.5±438.6c | 1441.4±440.9c | 8.43±0.74a | 0.99±0.01ab |
| S | 1968.7±546.6bc | 1969.1±545.8bc | 9.29±0.62a | 0.99±0.01a |
| PW | 1421.6±955.3a | 1403.3±922.7a | 6.95±2.18ab | 0.93±0.06bc |
| DW | 2834.2±900.4c | 2807.2±882.1c | 8.11±1.39b | 0.96±0.03c |
| R | 2351.3±74.3ab | 2268.6±53.4ab | 8.18±0.09ab | 0.96±0.01bc |
| Statistical significance is assessed by one-way ANOVA, P < 0.05. Different lowercase letters shows statistically significant difference. | ||||
表选项
群落组成上,放线菌门(Actinobacteria)、变形菌门(Proteobacteria)以及酸杆菌门(Acidobacteria)在微生物群落中占主导地位,相对丰度为52.09%-90.77% (图 2-A)。基于ASV进行非度量多维尺度(NMDS)分析,不同生境的微生物群落结构具有生境特异性(stress < 0.2,P < 0.001),尤其在液相样本中微生物群落差异更为突出,而固相样本相对液相样本样品则更为分散(图 2-C)。不同生境微生物群落的差异具体表现为:放线菌门(Actinobacteria)的相对丰度在岩壁样品中丰度较高;变形菌门(Proteobacteria)和拟杆菌门(Bacteroidetes)的相对丰度在液相样本样品中明显高于岩壁和沉积物;酸杆菌门(Acidobacteria)、芽单胞菌门(Gemmatimonadetes)和硝化螺旋菌门(Nitrospirae)在岩壁和沉积物中相对丰度高于液相样本。绿弯菌门(Chloroflexi)在沉积物样品中相对丰度较高(图 2-A)。在变形菌门中,β-变形菌纲(Betaproteobacteria,8.13%-30.66%)和γ-变形菌纲(Gammaproteobacteria,5.07%-42.65%)在所有样本中都具有较高的相对丰度。γ-变形菌纲(Gammaproteobacteria)在岩壁中相对丰度要高于同一位点的沉积物样本,而水潭积水、滴水与地下河河水中的γ-变形菌纲(Gammaproteobacteria)的相对丰度均较高(9.82%-42.65%) (图 2-B)。
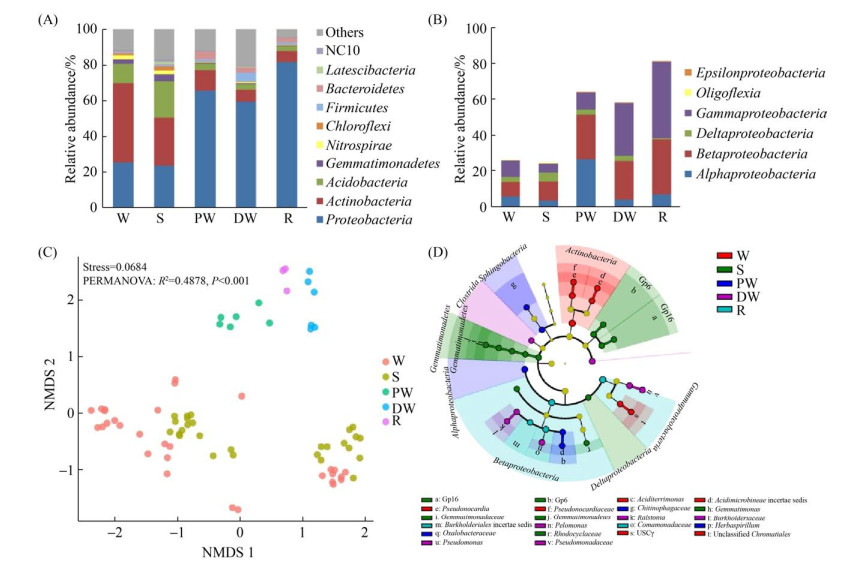 |
| 图 2 广西桂林罗汉肚洞微生物群落结构及不同生境中微生物群落的指示类群 Figure 2 Microbial community compositions and microbial indicators of different niches in the Luohandu cave, Guilin city, Guangxi province. A: the microbial composition of the top10 phyla. B: the relative abundances of the classes within Proteobacteria. C: non-metric multidimensional scaling analysis of microbial communities based on ASVs. D: the indictor groups in different niches based on LEfSe (Linear discriminant analysis Effect Size) with the LDA score > 4. |
| 图选项 |
LEfSe分析鉴定出24个指示类群,隶属于不同的生境(图 2-D)。沉积物的指示类群以酸杆菌门(Acidobacteria)和芽单胞菌门(Gemmatimonadetes)为主,岩壁则以放线菌门的酸微菌目(Acidimicrobiales)、假诺卡氏菌属(Pseudonocardia)和变形菌门(Actinobacteria)的USCγ类群为指示类群。液相样本的指示类群为变形菌门(Proteobacteria),滴水的指示类群还有厚壁菌门的梭菌纲(Clostrida),水潭积水的指示类群还包括拟杆菌门的噬几丁质杆菌科(Chitinophagaceae)。
2.3 微生物群落结构与环境因子的关系 微生物群落与环境因子之间的冗余分析(RDA)结果显示,第一轴和第二轴对固相样本微生物群落变化的解释度分别为70.94%和3.20% (图 3-A),其中,温度、SO42-、Cl-以及风化参数(Ca/Mg)对于微生物群落结构具有显著影响(P < 0.05),这些因子对群落差异的解释度分别为46.1%、25.5%、2.5%和2.5% (图 3-A)。温度和CH4对于固相样本中丰度前十的属产生显著影响(图 3-C)。液相样本中细菌群落结构与环境因子的冗余分析结果表明,溶解氧(DO)和电导率(EC)对微生物群落结构影响显著,这两个环境因子的解释度分别为47.3%和11.0% (图 3-B)。而液相样本中,电导率与不动杆菌属(Acinetobacter)显著负相关,pH与假单胞菌属(Pseudomonas)显著正相关,同时NO3-也是影响物种丰度的一个关键因子,与草螺菌属(Herbaspirillum)和振动单胞菌属(Vibrionimonas)呈显著正相关(图 3-D)。
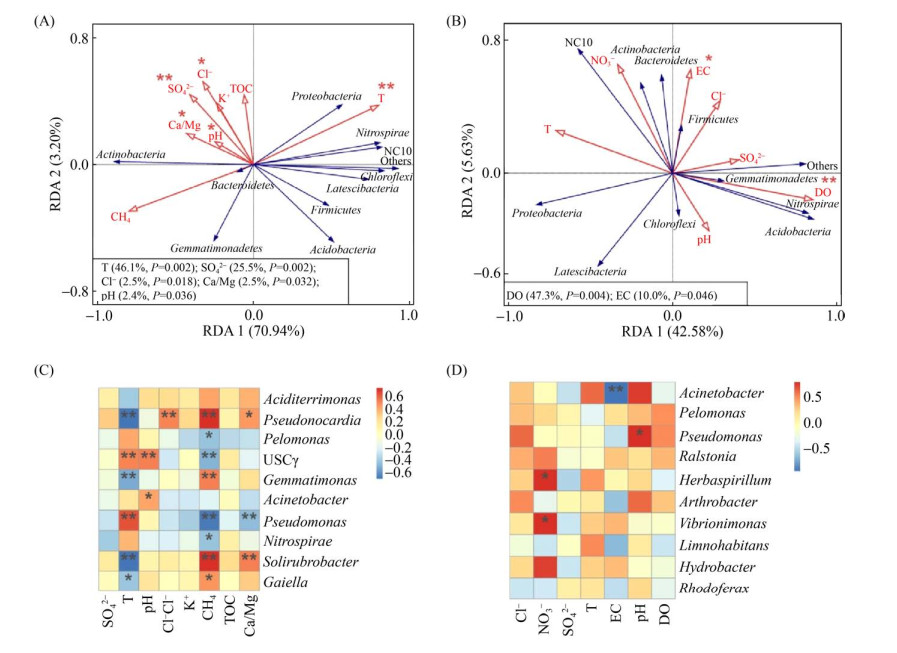 |
| 图 3 广西桂林罗汉肚洞固相样本(A)和液相样本(B)微生物群落组成及其与环境因子之间的RDA分析 Figure 3 Redundancy analysis (RDA) between environmental variables and microbial communities in solid samples (weathered rocks and sediments) (A) and in liquid samples (dripping water, pool water and ground river water samples) (B). The correlation between environmental factors and the top 10 microbial genera in solid samples (C) and in liquid samples (D). *: P < 0.05, **: P < 0.01, ***: P < 0.001. |
| 图选项 |
2.4 洞穴中微生物群落的共生网络 相关性网络包含309个节点以及3028条边。节点间以正相关(99.34%)为主,只有少数是负相关(0.66%) (图 4-A)。模块性分析发现在罗汉肚洞穴的微生物共现性网络中存在8个主要模块。网络的拓扑参数分别为:平均度21.60,平均路径3.40,网络直径9,聚集系数0.73,模块化指数0.63。网络中的节点被划分为12个细菌门,其中变形菌门、放线菌门、酸杆菌门3个门分布广泛,占全部节点的77%以上(图 4-B)。根据中介中心性(betweenness centrality)评分,丰度排名前三的节点被划分为关键类群,分别属于鞘氨醇单胞菌属(Sphingomonas)、拜叶林克氏菌科(Beijerinckiaceae)的一个未培养属和假诺卡氏菌属(Pseudonocardia)。
 |
| 图 4 广西桂林罗汉肚洞穴中微生物群落的共生网络(基于ASVs的Spearman相关性) Figure 4 Co-occurrence networks of the microbial communities in the Luohandu Cave (based on Spearman's correlation between ASVs) Guilin city, Guangxi province. All the connections have a correlation coefficient > |0.6| and a P < 0.05. Nodes were colored by modules (A) and by bacterial phylum (B). C: the relative abundances of ASVs in different niches within individual modules. The size of each node is proportional to the number of connections. Positive links are in and negative one in green. |
| 图选项 |
网络中所有节点被分成8个主要模块(图 4-A),1-6模块占所有节点的多数。各个模块中的节点表现出对于生境的不同偏好。如图 4-C所示,在模块1和2中,多数节点来自岩壁;在模块3、4和6中,多数节点则来自于沉积物;液相样本的节点多出现在模块5中。
2.5 固相样品和液相样本的群落构建 罗汉肚洞穴微生物群落的βNTI值大多位于-2到+2之间(图 5-A),表明微生物群落构建以随机性过程占主导地位,随机过程在固相样本和液相样本群落构建中的贡献率分别为51.25%和64.76% (图 5-B)。具体到单一的生态学过程,固相样本中确定性过程中的异质选择和均质选择分别贡献26.75%和21.99%;随机性过程中的均质扩散和不明确过程分别贡献40.42%和10.46%。液相样本中,贡献度由高到低依次为不明确过程(28.57%)、同质选择(25.71%)、均质扩散(24.76%),扩散限制(10.48%)和异质选择(9.52%) (图 5-B)。
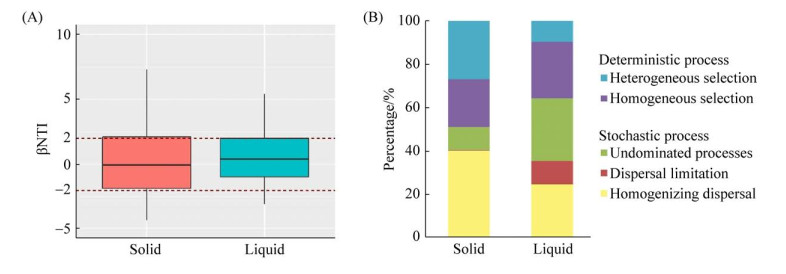 |
| 图 5 广西桂林罗汉肚固相样本和液相样本的βNTI (A)以及不同生态学过程对群落构建的贡献(B) Figure 5 βNTI values (A) and the contribution of individual ecological processes to community assembly (B) in solid and liquid samples in the Luohandu Cave, Guilin city, Guangxi province. |
| 图选项 |
3 讨论 3.1 洞穴微生物的生境特异性及其环境驱动因子 罗汉肚洞穴微生物群落显示出高度的生境特异性,不同生境优势类群不同。前人的研究表明岩壁微生物群落以放线菌占主导地位[35-36],本研究的结果也证明了这一观点。而沉积物中放线菌、变形菌以及酸杆菌门丰度都相对较高,这在碱性土壤中较为常见[16],同时这些类群在前人对于沉积物的研究中也得到了证实[16]。已有研究表明,β-变形菌纲(Betaproteobacteria)是岩溶水潭积水小生境中的核心微生物,广泛存在于不同洞穴的水潭积水中,并占据主导地位[37-38],这与本研究中水潭积水的群落特征相一致。而在滴水小生境中,则是γ-变形菌纲(Gammaproteobacteria)占主导地位[18]。以上结果均表明洞穴微生物对小生境的特异性具有广泛性,并且洞穴中液体样本与固体样本呈现出显著差异。在不同的小生境中指示类群表现出明显差异,在岩壁中酸土单孢菌属(Aciditerrimonas)和假诺卡氏菌属(Pseudonocardia)以及USCγ为指示类群。酸土单孢菌属(Aciditerrimonas)常出现于土壤环境中,与铁的氧化还原相关[39-40]。有文献表明USCγ在风化岩壁中占主导地位,对低浓度甲烷具有较高的亲和力,具有氧化大气中痕量甲烷的作用[12]。在以往研究中假诺卡氏菌属(Pseudonocardia)也被鉴定为岩壁生境的指示类群[41],并且是旧石器时代壁画上微生物群落的主要组成部分和具有活性的关键类群[42-43]。在滴水中,指示类群为假单胞菌属(Pseudomonas),多报道于根际土壤,且往往具有固氮基因[44-45]。在水潭积水中的指示类群为噬几丁质杆菌科(Chitinophagaceae),该科常被发现于淡水湖泊和水库等水体环境中[46-47],是甲烷氧化以及反硝化作用的重要组成部分[48],在这一科中被报道的许多属均具有还原硝酸盐的能力[49-50]。而草螺菌(Herbaspirillum)常出现于根际,是一种固氮细菌,与植物的生长息息相关[51]。液相样本中的指示类群与氮循环密切有关,如固氮作用和生物脱氮相关,同根际土壤的微生物类似,这可能与水流或滴水的携带外来微生物进入洞穴有关,这些微生物在洞穴氮循环中的作用还有待深入研究。洞穴水生小生境的群落结构相似,可能与较长的水岩相互作用导致水体样本的理化参数趋于相似,从而形成相对稳定的水生小生境有关。
SO42-和温度对于固相样本微生物群落结构影响显著。有研究表明,SO42-是影响细菌群落结构的主要影响因素,并且会对放线菌门中的异养类群产生影响[52]。在6、7号点位微生物群落结构的变化可能与温度显著相关,从6号位点的10 ℃升至7号位点的14 ℃,这一显著的温度跃升(4 ℃)影响了群落组成,导致6号点之前的微生物群落与7号点之后的微生物群落产生了显著的差异(图 2-C)。关于全球变暖的原位增温实验的研究中,在两年甚至更长的时间增温4 ℃,土壤的碳损失增加37%-55%,对于微生物活性以及呼吸作用也产生强烈影响[53-54]。洞穴中甲烷氧化微生物也可能受温度影响,有研究表明,温度会影响甲烷的氧化,在低于30 ℃时,甲烷氧化速率会随温度的升高而加快[55]。同时,温度也会在一定程度上影响微生物群落结构的变化[56]。本文中也发现,温度与USCγ类群丰度呈显著正相关(图 3-C)。温度同时也是影响固氮微生物群落的重要因素[57-58],洞穴中温度与假单胞菌属(Pseudomonas)呈显著正相关(图 3-C),同时也会对革兰氏阳性菌和放线菌的丰度产生影响[59]。本研究结果还发现,温度与洞穴微生物群落的结构和多样性显著相关,且温度与CH4浓度对群落物种影响相反(图 3-C),证明温度可能与甲烷消耗呈正相关性[60]。
pH常被认为是影响土壤微生物群落的重要因素。对北美和南美土壤微生物的研究发现,放线菌门(Actinobacteria)随着pH的增高而增加,而酸杆菌门(Acidobacteria)则随pH增高而减少[61]。同时利用宏基因组中对于β-葡萄糖苷酶基因序列的结果分析,在pH为5时酸杆菌门丰度增加,而在pH为7时放线菌的相对丰度增加[62]。在对湖北和尚洞的研究中,pH对细菌群落结构也具有显著影响,且酸杆菌门(Acidobacteria)与pH同样呈显著负相关,但放线菌门(Actinobacteria)却并未发现有显著相关性[16]。在本研究中,pH也显著影响洞穴固相样本中的微生物群落,与酸杆菌门(Acidobacteria)显著负相关[21],且与USCγ呈显著正相关。USCγ通常被发现于碱性环境中[63-64],这与本研究中所有样本pH呈碱性的结果吻合。风化程度不同会对岩石以及土壤中微生物群落和生物多样性产生显著影响[65],从而对生态系统功能产生间接影响[19]。
而在液相样品中影响细菌群落结构的环境因子则全然不同。在前人研究中,湖水中细菌群落与溶解氧(DO)显著相关[66-67],与本研究中结果一致。而电导率在本研究中也对液相样本产生显著的影响(图 3-D),可能是由于本研究中液相样本来自3个不同的小生境分别为水潭积水、滴水和地下河河水,其水力停留时间不同所导致的。
3.2 洞穴中微生物共生网络中的关键类群及其潜在的生态功能 通过共生网络可以揭示部分发生在微生物群落中的复杂的相互作用。洞穴微生物群落呈现较好的模块性(模块化指数大于0.4),这表明微生物群落中存在稳定复杂的相互作用和强大的模块化结构。罗汉肚洞穴中微生物群落聚集系数高,说明微生物群落的共生网络存在一个强相关性[68]。共现指数越低、聚类系数越大、负边比例越低,说明微生物群落具有较高的协同性,以往的报道大都显示生态网络的连接度较低[69],这在罗汉肚洞穴的微生物共生网络中也得到了验证。不同的微生物类群之间以正相关关系为主,这在其他洞穴以及其他类型的深地生物圈中也有印证[70]。这可能表明洞穴中的微生物群落因生态位的分异,缺少竞争,因此不同的微生物类群间以协作的关系为主[71-72]。
前人研究表明,较高的中介中心性评分对于维持生态网络的连通性十分重要[73]。鞘氨醇单胞菌属(Sphingomonas)、拜叶林克氏菌科(Beijerinckiaceae)的一个未培养属和假诺卡氏菌属(Pseudonocardia)在罗汉肚洞穴共生网络中的中介中心性评分位列前三。因此,推测它们可能在维持溶洞群落的结构和功能方面起着重要作用。鞘氨醇单胞菌属(Sphingomonas)对于多环芳烃具有极为广泛的代谢能力,广泛分布于海水[74]、淡水[75]、地下水[76]、污水[77]、沉积物[78]和陆地土壤[79]中。利用多环芳烃和各种代谢中间体作为碳源生长,并将它们转化成二氧化碳以获取生长的能源,是碳循环中重要的一环。拜叶林克氏菌科(Beijerinckiaceae)是好氧或兼性厌氧菌,在好氧或微氧条件下可以固定大气中的氮,有研究发现拜叶林克氏菌科(Beijerinckiaceae)作为与大气甲烷氧化菌USCα同源的甲烷氧化菌,生长过程中会转录较多的nifH基因,可能会在氮固定方面起主导作用[80]。假诺卡氏菌属(Pseudonocardia)主要与纤维素降解、抗生素合成有关[81]。现有研究表明,假诺卡氏菌属中一些分离出的菌株表现出固氮作用,可以进行异养氨氧化[82],同时也还有一些菌株可以用醚类、醇类、苯类或四氢呋喃作为碳源[83]。这些可以利用环境中难以降解的化合物作为碳源的菌属,在细菌群落中可能起到连接作用,对生态系统的维持起到重要作用。因此,本研究发现的关键类群可能会在洞穴生态系统的碳循环、氮循环中发挥重要作用。
3.3 群落构建的生态学过程 群落构建被广泛应用于研究影响微生物群落的生态学过程,对建立微生物群落的组成和生态功能之间的联系至关重要[84-87]。总体而言,影响微生物群落的生态学过程可以分为确定性过程和随机性过程,前者包括异质选择和均质选择,后者包括均质扩散、扩散限制和不明确过程[88-89]。不同的生态学过程对微生物群落的影响不同,均质选择最终会导致微生物群落的趋同,而异质选择则会驱使微生物群落产生差异。随机性过程中的均质扩散会导致微生物群落组成上的相似性提高,这主要是由于高扩散速率造成的;相反如果扩散受限,微生物群落的相似性则会降低。
本研究的结果显示,罗汉肚洞穴固相样本中确定性过程和随机性过程对群落构建的贡献基本相等,但就单一的生态学过程来讲,随机性过程中的均质扩散贡献最大(40.42%),表明微生物在固相样本中具有相对较高的扩散速率;异质选择对固相样本的群落构建的贡献位居第二(26.75%),这与不同固相样本间的环境因子的差异有关,可以发现随着确定性过程贡献的增加,环境因子(如温度、pH、Ca/Mg等)对于微生物群落的影响更为重要,主要环境因子对群落解释度(74.14%)远高于液体样本(48.21%) (图 3-A,B)。如随着采样位点距洞口距离的加大,采样点的温度会逐渐升高,这对微生物群落的组成及某些类群的相对丰度都会造成重要的影响(图 3-A,C),因此会引起微生物群落之间的差异。在流动性较低的固相中,由于对有限资源的竞争,导致选择可能更加激烈。而在液相样本中,对群落构建贡献最大的前3个生态学过程分别是不明确过程(25.71%)、同质选择(24.76%)和均质扩散,随机性过程占主导。而同质选择和均质扩散的结果都是导致微生物群落的相似度增加,微生物群落呈现趋同的现象,这可能是由于水相样本具有良好的流动性,且水岩相互作用长,会使得样本本身的理化参数趋同和微生物的扩散速率增加,因此导致微生物群落的相似性增加(图 2-C),不同水体样本的微生物群落紧密聚集在一起(图 4-C)。
4 结论 桂林罗汉肚洞穴中微生物群落表现出强烈的生境特异性,岩壁样品以放线菌门(Actinobacteria)为优势类群,沉积物中的优势类群则为酸杆菌门(Acidobacteria),所有水样微生物群落均以γ-变形菌纲(Gammaproteobacteria)为主。固相样本和液相样本中的微生物群落结构的环境驱动机制不同,风化指数以及SO42-浓度显著影响罗汉肚岩壁和沉积物等固相样本中微生物的群落结构,而电导率(EC)和溶解氧(DO)则是显著影响滴水、水潭积水以及地下河河水等液相样本中的微生物群落。洞穴微生物的共生网络具有明显的模块性,模块显示出微生物对生境的偏好性,液相样本中的微生物多出现在同一模块中。洞穴细菌群落中的指示类群可能在碳氮生物地球化学循环中发挥关键作用。固相样本与液相样本中微生物群落构建的生态学过程差异明显,固相样本中群落构建确定性过程(48.75%)与随机性过程(51.25%)的贡献基本相等,但液相样本中微生物的群落构建由于液体的强流动性则以随机过程占主导(64.76%)。本研究结果不仅确定了洞穴中各个小生境微生物群落的差异,而且清晰地回答了洞穴环境中不同相态的样本中微生物群落的环境驱动机制及其群落构建的生态学过程,对深刻认识陆地深部生物圈具有重要意义。
References
| [1] | Engel AS. Microbial diversity of cave ecosystems. Geomicrobiology: Molecular and Environmental Perspective. Dordrecht: Springer Netherlands, 2010: 219-238. |
| [2] | Legatzki A, Ortiz M, Neilson JW, Dominguez S, Andersen GL, Toomey RS, Pryor BM, Pierson LSⅢ, Maier RM. Bacterial and archaeal community structure of two adjacent calcite speleothems in Kartchner Caverns, Arizona, USA. Geomicrobiology Journal, 2011, 28(2): 99-117. DOI:10.1080/01490451003738465 |
| [3] | Huang QH, Cai YL, Xing XS. Rocky desertification, antidesertification, and sustainable development in the karst mountain region of southwest China. Ambio, 2008, 37(5): 390-392. DOI:10.1579/08-S-493.1 |
| [4] | Tomczyk-?ak K, Zielenkiewicz U. Microbial diversity in caves. Geomicrobiology Journal, 2016, 33(1): 20-38. DOI:10.1080/01490451.2014.1003341 |
| [5] | Barton H, Jurado V. What's up down there? Microbial diversity in caves microorganisms in caves survive under nutrient-poor conditions and are metabolically versatile and unexpectedly diverse. Microbe, 2007, 2(3). |
| [6] | Anbu P, Kang CH, Shin YJ, So JS. Formations of calcium carbonate minerals by bacteria and its multiple applications. SpringerPlus, 2016, 5(1): 1-26. DOI:10.1186/s40064-015-1659-2 |
| [7] | Al Qabany A, Soga K, Santamarina C. Factors affecting efficiency of microbially induced calcite precipitation. Journal of Geotechnical and Geoenvironmental Engineering, 2012, 138(8): 992-1001. DOI:10.1061/(ASCE)GT.1943-5606.0000666 |
| [8] | Lian B, Chen Y, Zhu LJ, Yang RD. Effect of microbial weathering on carbonate rocks. Earth Science Frontiers, 2008, 15(6): 90-99. DOI:10.1016/S1872-5791(09)60009-9 |
| [9] | De Mandal S, Chatterjee R, Kumar NS. Dominant bacterial Phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiology, 2017, 17(1): 1-9. DOI:10.1186/s12866-016-0921-2 |
| [10] | Zhao R, Wang HM, Yang H, Yun Y, Barton HA. Ammonia-oxidizing Archaea dominate ammonia-oxidizing communities within alkaline cave sediments. Geomicrobiology Journal, 2017, 34(6): 511-523. DOI:10.1080/01490451.2016.1225861 |
| [11] | Webster KD, Drobniak A, Etiope G, Mastalerz M, Sauer PE, Schimmelmann A. Subterranean karst environments as a global sink for atmospheric methane. Earth and Planetary Science Letters, 2018, 485: 9-18. DOI:10.1016/j.epsl.2017.12.025 |
| [12] | Zhao R, Wang HM, Cheng XY, Yun Y, Qiu X. Upland soil cluster γ dominates the methanotroph communities in the karst Heshang Cave. FEMS Microbiology Ecology, 2018, 94(12): 826-836. |
| [13] | Cheng XY, Liu XY, Wang HM, Su CT, Zhao R, Bodelier PLE, Wang WQ, Ma LY, Lu XL. USCγ dominated community composition and cooccurrence network of methanotrophs and bacteria in subterranean karst caves. Microbioogyl Spectrum, 2021, 9(1): e00820-21. |
| [14] | Zhu HZ, Zhang ZF, Zhou N, Jiang CY, Wang BJ, Cai L, Wang HM, Liu SJ. Bacteria and metabolic potential in karst caves revealed by intensive bacterial cultivation and genome assembly. Applied and Environmental Microbiology, 2021, 87(12): e0057721-21. DOI:10.1128/aem.00577-21 |
| [15] | Bartram AK, Jiang XP, Lynch MDJ, Masella AP, Nicol GW, Dushoff J, Neufeld JD. Exploring links between pH and bacterial community composition in soils from the Craibstone experimental farm. FEMS Microbiology Ecology, 2014, 87(2): 403-415. DOI:10.1111/1574-6941.12231 |
| [16] | Yun Y, Wang HM, Man BY, Xiang X, Zhou JP, Qiu X, Duan Y, Engel AS. The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification. Frontiers in Microbiology, 2016, 7: 1955-1971. |
| [17] | Walker TWN, Kaiser C, Strasser F, Herbold CW, Leblans NIW, Woebken D, Janssens IA, Sigurdsson BD, Richter A. Microbial temperature sensitivity and biomass change explain soil carbon loss with warming. Nature Climate Change, 2018, 8(10): 885-889. DOI:10.1038/s41558-018-0259-x |
| [18] | Yun Y, Cheng XY, Wang WQ, Wang HM. Seasonal variation of bacterial community and their functional diversity in drip water from a karst cave. Chinese Science Bulletin, 2018, 63(36): 3932-3944. (in Chinese) 云媛, 程晓钰, 王纬琦, 王红梅. 喀斯特洞穴滴水细菌群落组成及其代谢功能的季节性变化. 科学通报, 2018, 63(36): 3932-3944. |
| [19] | Hu A, Wang JJ, Sun H, Niu B, Si GC, Wang J, Yeh CF, Zhu XX, Lu XC, Zhou JZ, Yang YP, Ren ML, Hu YL, Dong HL, Zhang GX. Mountain biodiversity and ecosystem functions: interplay between geology and contemporary environments. The ISME Journal, 2020, 14(4): 931-944. DOI:10.1038/s41396-019-0574-x |
| [20] | Cheng XY, Yun Y, Wang HM, Ma LY, Tian W, Man BY, LiuCY. Contrasting bacterial communities and their assembly processes in karst soils under different land use. Science of the Total Environment, 2021, 751: 142263. DOI:10.1016/j.scitotenv.2020.142263 |
| [21] | Shen CC, Xiong JB, Zhang HY, Feng YZ, Lin XG, Li XY, Liang WJ, Chu HY. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biology and Biochemistry, 2013, 57: 204-211. DOI:10.1016/j.soilbio.2012.07.013 |
| [22] | Jiang HC, Dong HL, Yu BS, Liu XQ, Li YL, Ji SS, Zhang CL. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan Plateau. Environmental Microbiology, 2007, 9(10): 2603-2621. DOI:10.1111/j.1462-2920.2007.01377.x |
| [23] | Tang Y, Liu YC, Yang J, Jiang HC. Gene diversity involved in kalvin pathway of carbon fixation and its response to environmental variables in surface sediments of the northern Qinghai-Tibetan Plateau lakes. Earth Science, 2018, 43(S1): 19-30. (in Chinese) 唐阳, 刘永超, 杨渐, 蒋宏忱. 青藏高原北部湖泊表层沉积物参与卡尔文循环的固碳基因多样性及其影响因素. 地球科学, 2018, 43(S1): 19-30. |
| [24] | Angly FE, Heath C, Morgan TC, Tonin H, Rich V, Schaffelke B, Bourne DG, Tyson GW. Marine microbial communities of the Great Barrier Reef lagoon are influenced by riverine floodwaters and seasonal weather events. PeerJournal, 2016, 4: e1511. DOI:10.7717/peerj.1511 |
| [25] | Yanagawa K, Morono Y, de Beer D, Haeckel M, Sunamura M, Futagami T, Hoshino T, Terada T, Nakamura KI, Urabe T, Rehder G, Boetius A, Inagaki F. Metabolically active microbial communities in marine sediment under high-CO2 and low-pH extremes. The ISME Journal, 2013, 7(3): 555-567. DOI:10.1038/ismej.2012.124 |
| [26] | Graw MF, D'Angelo G, Borchers M, Thurber AR, Johnson JE, Zhang CL, Liu HD, Colwell FS. Energy gradients structure microbial communities across sediment horizons in deep marine sediments of the South China Sea. Frontiers in Microbiology, 2018, 9: 729. DOI:10.3389/fmicb.2018.00729 |
| [27] | Tian W, Wang HM, Xiang X, Wang RC, Xu Y. Structural variations of bacterial community driven by Sphagnum microhabitat differentiation in a subalpine peatland. Frontiers in Microbiology, 2019, 10: 1661. DOI:10.3389/fmicb.2019.01661 |
| [28] | Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics, 2008, 4(11): e1000255. DOI:10.1371/journal.pgen.1000255 |
| [29] | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS, 2011, 108(Supplement_1): 4516-4522. DOI:10.1073/pnas.1000080107 |
| [30] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pe?a AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QⅡME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [31] | Cai YF, Zhou X, Shi LM, Jia ZJ. Atmospheric methane oxidizers are dominated by upland soil cluster alpha in 20 forest soils of China. Microbial Ecology, 2020, 80(4): 859-871. DOI:10.1007/s00248-020-01570-1 |
| [32] | Stegen JC, Lin XJ, Konopka AE, Fredrickson JK. Stochastic and deterministic assembly processes in subsurface microbial communities. The ISME Journal, 2012, 6(9): 1653-1664. DOI:10.1038/ismej.2012.22 |
| [33] | Stegen JC, Lin XJ, Fredrickson JK, Chen XY, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. Quantifying community assembly processes and identifying features that impose them. The ISME Journal, 2013, 7(11): 2069-2079. DOI:10.1038/ismej.2013.93 |
| [34] | Webb CO. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. The American Naturalist, 2000, 156(2): 145-155. DOI:10.1086/303378 |
| [35] | Rangseekaew P, Pathom-Aree W. Cave Actinobacteria as producers of bioactive metabolites. Frontiers in Microbiology, 2019, 10: 387. DOI:10.3389/fmicb.2019.00387 |
| [36] | Pa?i? L, Kov?e B, Sket B, Herzog-Velikonja B. Diversity of microbial communities colonizing the walls of a karstic cave in Slovenia. FEMS Microbiology Ecology, 2010, 71(1): 50-60. DOI:10.1111/j.1574-6941.2009.00789.x |
| [37] | Shabarova T, Pernthaler J. Karst pools in subsurface environments: collectors of microbial diversity or temporary residence between habitat types. Environmental Microbiology, 2010, 12(4): 1061-1074. DOI:10.1111/j.1462-2920.2009.02151.x |
| [38] | Shabarova T, Widmer F, Pernthaler J. Mass effects meet species sorting: transformations of microbial assemblages in epiphreatic subsurface karst water pools. Environmental Microbiology, 2013, 15(9): 2476-2488. DOI:10.1111/1462-2920.12124 |
| [39] | Itoh T, Yamanoi K, Kudo T, Ohkuma M, Takashina T. Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic Actinobacterium isolated from a solfataric field. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(6): 1281-1285. DOI:10.1099/ijs.0.023044-0 |
| [40] | Huang S, Jaffé PR. Isolation and characterization of an ammonium-oxidizing iron reducer: Acidimicrobiaceae sp. A6. PLoS One, 2018, 13(4): e0194007. DOI:10.1371/journal.pone.0194007 |
| [41] | Zhu HZ, Zhang ZF, Zhou N, Jiang CY, Wang BJ, Cai L, Liu SJ. Diversity, distribution and co-occurrence patterns of bacterial communities in a karst cave system. Frontiers in Microbiology, 2019, 10: 1726. DOI:10.3389/fmicb.2019.01726 |
| [42] | Stomeo F, Portillo MC, Gonzalez JM. Assessment of bacterial and fungal growth on natural substrates: consequences for preserving caves with prehistoric paintings. Current Microbiology, 2009, 59(3): 321-325. DOI:10.1007/s00284-009-9437-4 |
| [43] | Stomeo F, Portillo MC, Gonzalez JM, Laiz L, Saiz-Jimenez C. Pseudonocardia in white colonizations in two caves with Paleolithic paintings. International Biodeterioration & Biodegradation, 2008, 62(4): 483-486. |
| [44] | Kuklinsky-Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environmental Microbiology, 2004, 6(12): 1244-1251. DOI:10.1111/j.1462-2920.2004.00658.x |
| [45] | Gomila M, Bowien B, Falsen E, Moore ERB, Lalucat J. Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(11): 2629-2635. DOI:10.1099/ijs.0.65149-0 |
| [46] | Liu LM, Su JQ, Guo YY, Wilkinson DM, Liu ZW, Zhu YG, Yang J. Large-scale biogeographical patterns of bacterial antibiotic resistome in the waterbodies of China. Environment International, 2018, 117: 292-299. DOI:10.1016/j.envint.2018.05.023 |
| [47] | Orr RJS, Rombauts S, van de Peer Y, Shalchian-Tabrizi K. Draft genome sequences of two unclassified Chitinophagaceae bacteria, IBVUCB1 and IBVUCB2, isolated from environmental samples. Genome Announcements, 2017, 5(41). DOI:10.1128/genomea.00787-17 |
| [48] | Waki M, Yasuda T, Yokoyama H, Hanajima D, Ogino A, Suzuki K, Yamagishi T, Suwa Y, Tanaka Y. Nitrogen removal by co-occurring methane oxidation, denitrification, aerobic ammonium oxidation, and anammox. Applied Microbiology and Biotechnology, 2009, 84(5): 977-985. DOI:10.1007/s00253-009-2112-7 |
| [49] | K?mpfer P, Lodders N, Falsen E. Hydrotalea flava gen. nov., sp. nov., a new member of the Phylum Bacteroidetes and allocation of the genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferruginibacter, Filimonas and Hydrotalea to the family Chitinophagaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(3): 518-523. DOI:10.1099/ijs.0.023002-0 |
| [50] | Zhang L, Wang Y, Wei LF, Wang Y, Shen XH, Li SQ. Taibaiella smilacinae gen. nov., sp. nov., an endophytic member of the family Chitinophagaceae isolated from the stem of Smilacina Japonica, and emended description of Flavihumibacter petaseus. International Journal of Systematicand Evolutionary Microbiology, 2013, 63(Pt_10): 3769-3776. DOI:10.1099/ijs.0.051607-0 |
| [51] | Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends in Microbiology, 1998, 6(4): 139-144. DOI:10.1016/S0966-842X(98)01229-3 |
| [52] | Xu Z, Woodhouse JN, Te SH, Yew-Hoong Gin K, He YL, Xu C, Chen L. Seasonal variation in the bacterial community composition of a large estuarine reservoir and response to Cyanobacterial proliferation. Chemosphere, 2018, 202: 576-585. DOI:10.1016/j.chemosphere.2018.03.037 |
| [53] | Nottingham AT, Meir P, Velasquez E, Turner BL. Soil carbon loss by experimental warming in a tropical forest. Nature, 2020, 584(7820): 234-237. DOI:10.1038/s41586-020-2566-4 |
| [54] | Melillo JM, Frey SD, DeAngelis KM, Werner WJ, Bernard MJ, Bowles FP, Pold G, Knorr MA, Grandy AS. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science, 2017, 358(6359): 101-105. DOI:10.1126/science.aan2874 |
| [55] | Reddy KR, Rai RK, Green SJ, Chetri JK. Effect of temperature on methane oxidation and community composition in landfill cover soil. Journal of Industrial Microbiology & Biotechnology, 2019, 46(9): 1283-1295. |
| [56] | Wilms R, Sass H, K?pke B, Cypionka H, Engelen B. Methane and sulfate profiles within the subsurface of a tidal flat are reflected by the distribution of sulfate-reducing bacteria and methanogenic Archaea. FEMS Microbiology Ecology, 2007, 59(3): 611-621. DOI:10.1111/j.1574-6941.2006.00225.x |
| [57] | Sharp CE, Brady AL, Sharp GH, Grasby SE, Stott MB, Dunfield PF. Humboldt's spa: microbial diversity is controlled by temperature in geothermal environments. The ISME Journal, 2014, 8(6): 1166-1174. DOI:10.1038/ismej.2013.237 |
| [58] | Dang HY, Luan XW, Zhao JY, Li J. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane seep sediments of the Okhotsk Sea. Applied and Environmental Microbiology, 2009, 75(7): 2238-2245. DOI:10.1128/AEM.02556-08 |
| [59] | Frey SD, Drijber R, Smith H, Melillo J. Microbial biomass, functional capacity, and community structure after 12 years of soil warming. Soil Biology and Biochemistry, 2008, 40(11): 2904-2907. DOI:10.1016/j.soilbio.2008.07.020 |
| [60] | King GM, Adamsen APS. Effects of temperature on methane consumption in a forest soil and in pure cultures of the methanotroph Methylomonas rubra. Applied and Environmental Microbiology, 1992, 58(9): 2758-2763. DOI:10.1128/aem.58.9.2758-2763.1992 |
| [61] | Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 2009, 75(15): 5111-5120. DOI:10.1128/AEM.00335-09 |
| [62] | Puissant J, Jones B, Goodall T, Mang DN, Blaud A, Gweon HS, Malik A, Jones DL, Clark IM, Hirsch PR, Griffiths R. The pH optimum of soil exoenzymes adapt to long term changes in soil pH. Soil Biology and Biochemistry, 2019, 138: 107601. DOI:10.1016/j.soilbio.2019.107601 |
| [63] | Knief C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Frontiers in Microbiology, 2015, 6: 1346. |
| [64] | Tveit AT, Hestnes AG, Robinson SL, Schintlmeister A, Dedysh SN, Jehmlich N, von Bergen M, Herbold C, Wagner M, Richter A, Svenning MM. Widespread soil bacterium that oxidizes atmospheric methane. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(17): 8515-8524. DOI:10.1073/pnas.1817812116 |
| [65] | Xi J, Wei ML, Tang BK. Differences in weathering pattern, stress resistance and community structure of culturable rock-weathering bacteria between altered rocks and soils. RSC Advances, 2018, 8(26): 14201-14211. DOI:10.1039/C8RA01268G |
| [66] | Diao MH, Sinnige R, Kalbitz K, Huisman J, Muyzer G. Succession of bacterial communities in a seasonally stratified lake with an anoxic and sulfidic hypolimnion. Frontiers in Microbiology, 2017, 8: 2511. DOI:10.3389/fmicb.2017.02511 |
| [67] | Kurilkina MI, Zakharova YR, Galachyants YP, Petrova DP, Bukin YS, Domysheva VM, Blinov VV, Likhoshway YV. Bacterial community composition in the water column of the deepest freshwater Lake Baikal as determined by next-generation sequencing. FEMS Microbiology Ecology, 2016, 92(7): fiw094. DOI:10.1093/femsec/fiw094 |
| [68] | Xue L, Ren HD, Li S, Leng XH, Yao XH. Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Frontiers in Microbiology, 2017, 8: 2377. DOI:10.3389/fmicb.2017.02377 |
| [69] | Poisot T, Gravel D. When is an ecological network complex? Connectance drives degree distribution and emerging network properties. PeerJournal, 2014, 2: e251-e261. |
| [70] | Dong YY, Gao J, Wu QS, Ai YL, Huang Y, Wei WZ, Sun SY, Weng QB. Co-occurrence pattern and function prediction of bacterial community in karst cave. BMC Microbiology, 2020, 20(1): 1-13. DOI:10.1186/s12866-019-1672-7 |
| [71] | Eiler A, Heinrich F, Bertilsson S. Coherent dynamics and association networks among lake bacterioplankton taxa. The ISME Journal, 2012, 6(2): 330-342. DOI:10.1038/ismej.2011.113 |
| [72] | Faust K, Raes J. Microbial interactions: from networks to models. Nature Reviews Microbiology, 2012, 10(8): 538-550. DOI:10.1038/nrmicro2832 |
| [73] | Martín González AM, Dalsgaard B, Olesen JM. Centrality measures and the importance of generalist species in pollination networks. Ecological Complexity, 2010, 7(1): 36-43. DOI:10.1016/j.ecocom.2009.03.008 |
| [74] | Gilewicz M, Nadalig T, Budzinski H, Doumenq P, Michotey V, Bertrand JC. Isolation and characterization of a marine bacterium capable of utilizing 2-methylphenanthrene. Applied Microbiology and Biotechnology, 1997, 48(4): 528-533. DOI:10.1007/s002530051091 |
| [75] | Schweitzer B, Huber I, Amann R, Ludwig W, Simon M. α- and β- proteobacteria control the consumption and release of amino acids on lake snow aggregates. Applied and Environmental Microbiology, 2001, 67(2): 632-645. DOI:10.1128/AEM.67.2.632-645.2001 |
| [76] | Tiirola MA, M?nnist? MK, Puhakka JA, Kulomaa MS. Isolation and characterization of Novosphingobium sp. strain MT1, a dominant polychlorophenol-degrading strain in a groundwater bioremediation system. Applied and Environmental Microbiology, 2002, 68(1): 173-180. DOI:10.1128/AEM.68.1.173-180.2002 |
| [77] | Coughlin MF, Kinkle BK, Bishop PL. Degradation of azo dyes containing aminonaphthol by Sphingomonas sp. strain 1CX. Journal of Industrial Microbiology and Biotechnology, 1999, 23(4/5): 341-346. |
| [78] | Fredrickson JK, Balkwill DL, Drake GR, Romine MF, Ringelberg DB, White DC. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Applied and Environmental Microbiology, 1995, 61(5): 1917-1922. DOI:10.1128/aem.61.5.1917-1922.1995 |
| [79] | S?rensen SR, Ronen Z, Aamand J. Isolation from agricultural soil and characterization of a Sphingomonas sp. able to mineralize the phenylurea herbicide isoproturon. Applied and Environmental Microbiology, 2001, 67(12): 5403-5409. DOI:10.1128/AEM.67.12.5403-5409.2001 |
| [80] | Vile MA, Kelman Wieder R, ?ivkovi? T, Scott KD, Vitt DH, Hartsock JA, Iosue CL, Quinn JC, Petix M, Fillingim HM, Popma JMA, Dynarski KA, Jackman TR, Albright CM, Wykoff DD. N2-fixation by methanotrophs sustains carbon and nitrogen accumulation in pristine peatlands. Biogeochemistry, 2014, 121(2): 317-328. DOI:10.1007/s10533-014-0019-6 |
| [81] | Carr G, Derbyshire ER, Caldera E, Currie CR, Clardy J. Antibiotic and antimalarial quinones from fungus-growing ant-associated Pseudonocardia sp. Journal of Natural Products, 2012, 75(10): 1806-1809. DOI:10.1021/np300380t |
| [82] | Liu ZP, Jia SF, Yu ZM, Liu SJ. Solation and characteristics of a new heterotrophic ammonia-oxidizing bacterium. Environmental Pollution & Control, 2005, 27(5): 337-339, 316. (in Chinese) 刘志培, 贾省芬, 俞志明, 刘双江. 新型异养氨氧化菌的分离鉴定及氨氧化特性. 环境污染与防治, 2005, 27(5): 337-339, 316. DOI:10.3969/j.issn.1001-3865.2005.05.006 |
| [83] | Sales CM, Mahendra S, Grostern A, Parales RE, Goodwin LA, Woyke T, Nolan M, Lapidus A, Chertkov O, Ovchinnikova G, Sczyrba A, Alvarez-Cohen L. Genome sequence of the 1, 4-dioxane-degrading Pseudonocardia dioxanivoransStrain CB1190. Journal of Bacteriology, 2011, 193(17): 4549-4550. DOI:10.1128/JB.00415-11 |
| [84] | Ferrenberg S, O'Neill SP, Knelman JE, Todd B, Duggan S, Bradley D, Robinson T, Schmidt SK, Townsend AR, Williams MW, Cleveland CC, Melbourne BA, Jiang L, Nemergut DR. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. The ISME Journal, 2013, 7(6): 1102-1111. DOI:10.1038/ismej.2013.11 |
| [85] | Graham E, Crump A, Resch C, Fansler S, Arntzen E, Kennedy D, Fredrickson J, Stegen J. Coupling spatiotemporal community assembly processes to changes in microbial metabolism. Frontiers in Microbiology, 2016: 7. |
| [86] | Wang JJ, Shen J, Wu YC, Tu C, Soininen J, Stegen JC, He JZ, Liu XQ, Zhang L, Zhang EL. Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. The ISME Journal, 2013, 7(7): 1310-1321. DOI:10.1038/ismej.2013.30 |
| [87] | Graham E, Stegen J. Dispersal-based microbial community assembly decreases biogeochemical function. Processes, 2017, 5(4): 65. DOI:10.3390/pr5040065 |
| [88] | Stegen JC, Lin XJ, Fredrickson JK, Konopka AE. Estimating and mapping ecological processes influencing microbial community assembly. Frontiers in Microbiology, 2015, 6: 370. |
| [89] | Vellend M. Conceptual synthesis in community ecology. The Quarterly Review of Biology, 2010, 85(2): 183-206. DOI:10.1086/652373 |
