王天乐, 刘喜朋


上海交通大学生命科学技术学院, 微生物代谢国家重点实验室, 上海 200240
收稿日期:2020-03-20;修回日期:2020-05-22;网络出版日期:2020-07-08
基金项目:国家自然科学基金(U1832161)
*通信作者:刘喜朋. Tel:+86-21-34204378; E-mail:xpliu@sjtu.edu.cn.
摘要:[目的] 克隆表达嗜热古菌Archaeoglobus fulgidus(A.fulgidus)来源的RecJ核酸酶基因(ORF编号AF_0699,NCBI数据库基因登陆号为AF_RS03550),对该重组蛋白的核酸酶活性及酶学特征进行鉴定和分析。[方法] 将A.fulgidus RecJ(AfuRecJ)核酸酶在大肠杆菌中进行重组表达,经亲和层析纯化得到电泳纯蛋白;利用人工合成的带有末端荧光标记的寡核苷酸作为底物,体外反应后,利用8 mol/L尿素变性聚丙烯酰胺凝胶电泳鉴定AfuRecJ核酸酶的水解产物。[结果] AfuRecJ核酸酶具有单链DNA特异性的3'-5'外切核酸酶活性,酶活性依赖于二价金属离子Mn2+或Mg2+,且Mn2+存在条件下的催化效率明显优于Mg2+;其最适反应温度范围为55-65℃;高于200 mmol/L的NaCl会显著抑制AfuRecJ的核酸酶活性。AfuRecJ还具有单链RNA 3'-5'外切酶活性,且活性高于单链DNA底物。单链核酸3'末端的磷酸基团对水解活性有一定抑制作用。AfuRecJ对单链核酸的长度有一定的选择性,可以有效水解长度≥4个核苷酸长度的单链RNA、≥12个核苷酸长度的单链DNA,而且对双链DNA中的3'单链DNA结构(3'突出单链尾巴与末端分叉结构)具有类似单链DNA的水解活性。[结论] 本研究证实AfuRecJ是一种单链核酸特异性的3'-5'外切核酸酶,且相比单链DNA,单链RNA为优势底物,推测其在胞内可能参与RNA降解与DNA修复。
关键词:嗜热古菌DNA修复RecJ核酸酶核酸酶
Expression, purification and characterization of RecJ nuclease from Archaeoglobus fulgidus
Tianle Wang, Xipeng Liu


State Key Laboratory of Microbial Metabolism, School of Life Science and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China
Received: 20 March 2020; Revised: 22 May 2020; Published online: 8 July 2020
*Corresponding author: Liu Xipeng. Tel:+86-21-34204378; E-mail:xpliu@sjtu.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (U1832161)
Abstract: [Objective] To clone, express and purify the RecJ nuclease gene (AF_0699) from Archaeoglobus fulgidus, identify and analyze its nuclease activity. [Methods] Recombinant RecJ nuclease (AfuRecJ) was expressed in E. coli and purified by affinity chromatography. The enzymatic properties of AfuRecJ nuclease were characterized in vitro using synthesized oligo (deoxy) nucleotides as substrate. [Results] AfuRecJ nuclease has a single-stranded DNA specific 3'–5' exonuclease activity, which depends on the divalent metal ions Mn2+ or Mg2+, and the catalytic efficiency of Mn2+ is significantly higher than that of Mg2+. The optimal reaction temperature is between 55 and 65 ℃. The presence of high concentration of NaCl decreases the exonuclease activity of AfuRecJ, and cleavage percentage greatly reduces at 200 mmol/L of NaCl. AfuRecJ shows 3'–5' exonuclease activity on single-stranded RNA, higher than that of single-stranded DNA. The 3' terminal phosphate group inhibits the AfuRecJ nuclease. AfuRecJ has different preferences for substrate length. It can effectively hydrolyze ssRNA≥4 nt, and ssDNA≥12 nt. Moreover, the hydrolysis activity of AfuRecJ on dsDNA with special structure (3' overhang and 3' fork) is similar to that of ssDNA. [Conclusion] The nuclease activity of AfuRecJ depends on the Mn2+ and can cleave ssDNA and ssRNA with a preference for RNA substrate.
Keywords: thermophilic archaeaDNA repairRecJ nucleasenuclease
核酸酶包括核酸内切酶和核酸外切酶,水解磷酸二酯键,参与细胞内DNA复制、重组、修复以及RNA成熟、加工等重要的代谢过程[1–2]。DHH磷酸酯水解酶(DHH超家族蛋白)是一大类能够水解磷酸酯键的酶的总称,参与多种核酸代谢过程。DHH磷酸酯水解酶不同亚家族间的氨基酸序列保守性很低,但均具有一个由3个连续氨基酸残基DHH (即天冬氨酸-组氨酸-组氨酸)构成的保守基序(motif),故统称为DHH超家族蛋白。DHH超家族包含许多水解磷酸二酯键的蛋白,根据结构域的差别可以划分为3个亚家族:Ⅰ型、Ⅱ型、Ⅲ型[2–3]。
RecJ蛋白最早发现于细菌同源重组过程[4],属于DHH超家族的Ⅰ型亚家族。细菌RecJ蛋白具有单链DNA特异性的5?–3?外切酶活性[5–7],因此被命名为RecJ核酸酶。RecJ核酸酶广泛存在于细菌。最近发现部分古菌编码一个或多个细菌RecJ核酸酶同源蛋白,例如詹氏甲烷球菌Methanocaldococcus jannaschii (M. jannaschii)编码3个RecJ同源蛋白[8]。但古菌RecJ同源蛋白在结构域组成上与细菌的RecJ核酸酶明显不同[5, 9],而与真核来源的Cdc45蛋白更为相似[10]。目前对细菌Deinococcus radiodurans (D. radiodurans)[11]、Thermus thermophilus (T. thermophilus)[5, 9]和古菌Pyrococcus furiosus (P. furiosus)[12],Thermococcus kodarensis (T. kodarensis)[13]等原核生物的RecJ核酸酶的生化功能和三级结构的认识较为清楚。
A. fulgidus是一种极端嗜热古菌,生长温度范围为60–95 ℃,最适生长温度为85 ℃,全基因组序列由美国马里兰基因研究所于1997年完成测定[14]。生物信息学分析表明A. fulgidus编码RecJ核酸酶的3个同源蛋白。经初步的活性测定,仅编号为AF_0699的RecJ蛋白显示出体外的核酸酶活性。而AF_0735本身在大肠杆菌中主要以包涵体形式表达,仅纯化到极少量可溶性蛋白,未检测到明显的核酸酶活性;同时AF_0075也未显示出明显的核酸酶活性,且AF_0075和其他两个RecJ蛋白的序列相似性较低。因此,为进一步研究古菌中的RecJ核酸酶的生物功能,本文首先选取A. fulgidus菌株编码的一个RecJ核酸酶(AF_0699),对其体外核酸酶活性进行了核酸酶活性测定和酶学特性表征。
1 材料和方法 1.1 材料 本实验所用的A. fulgidus基因组DNA源自美国ATCC。大肠杆菌菌株DH5α、Rosetta (DE3)感受态细胞购自北京全式金公司。用于构建表达载体的引物和测定核酸酶活性的损伤寡核苷酸底物由生工生物工程(上海)股份有限公司合成,底物序列见表 1。本实验中所用到的PrimeSTAR DNA聚合酶、Nde I和Hind III限制性内切酶、DNA Ladder、蛋白Marker购自TaKaRa公司,ClonExpress II一步克隆试剂盒购自诺唯赞公司。PCR产物纯化和胶回收试剂盒、质粒提取试剂盒、Bradford蛋白浓度测定试剂盒购自生工生物工程(上海)股份有限公司。Ni-NTA蛋白纯化树脂为Bio-Rad公司产品。
表 1. 分析AfuRecJ核酸酶活性的寡核苷酸序列 Table 1. Oligonucleotides and oligodeoxynucleotides used for analyzing AfuRecJ nuclease activity
| Number | Sequences (5?→3?) | Figures |
| DD286 | *TCCGATAGCCAGATATCTTGACA | 1,2 |
| DD286 JL510 | *TCCGATAGCCAGATATCTTGACA *uccgauagccagauaucuugacu | 3 |
| DD305 DD306 DD264 DD265 | *TCCGATAGCCAGATATCTTGACA TCCGATAGCCAGATATCTTGACA* *csasgscscsasgsgsusgsuscsuscsascsu asgscscsgsascsuscsgscscsascsasgsu* | 4 |
| DD286 DD297 DD247 DD256 | *TCCGATAGCCAGATATCTTGACA *TCCGATAGCCAGATATCTTGACAp *cggagaugacgg *cggagaugacggp | 5 |
| DD282 DD283 DD284 DD285 DD286 DD287 DD288 DD289 | *CGAT *TCCGAT *TCCGATAGCCAG *TCCGATAGCCAGATATC *TCCGATAGCCAGATATCTTGACA *TCCGATAGCCAGATATCTTGTGAGCGTGGG *AGGCTGCGGTCGAGTTGACAGCACTGCACGCATTACTGAGCT *CTCCAGTGGTGTTCGGCTCCGATAGCCAGATATCTTGTGACGTGACGTG CGTAATGAC | 6 |
| DD245 DD246 DD247 DD248 | *acgu *ugacgu *cggagaugacgg *cgagcggagaugacgg | |
| DD288 JL1196 JL390 JL391 JL392 | *AGGCTGCGGTCGAGTTGACAGCACTGCACGCATTACTGAGCT AGCTCAGTAATGCGTGCAGTGCTGTCAACTCGACCGCAGCCT TCGAGTCATTACGCTGCAGTGCTGTCAACTCGACCGCAGCCT TGCAGTGCTGTCAACTCGACCGCAGCCT TTTTTAGCTCAGTAATGCGTGCAGTGCTGTCAACTCGACCGCAGCCT | 7 |
| *: fluorescent group FAM; s: phosphorothioate modification; p: phosphorylation modification; Uppercase: DNA base; Lowercase: RNA base. | ||
表选项
1.2 表达载体构建 以A. fulgidus基因组DNA作为模板,利用PrimeSTAR DNA聚合酶PCR扩增recj基因。正向引物序列为5?-TGCCGCGCGGCAGCCATATG AGTTCGGGCTCGGCAAT-3?,反向引物序列为5?-TCGAGTGCGGCCGCAAGCTTCTAAGCGAGCATGGCACAGC-3? (带下划线斜体碱基表示酶切位点)。PCR扩增条件为95 ℃ 5 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 2 min。PCR产物经纯化试剂盒纯化。用Nde I和Hind III消化pET28a质粒,将其线性化,作为重组法基因克隆的质粒底物。用一步重组克隆试剂盒连接recj基因片段和线性化的pET28a质粒,取10 μL反应产物转化大肠杆菌DH5α,挑取单克隆培养,菌落PCR鉴定阳性克隆,并进行DNA序列测定,确定AfuRecJ核酸酶的基因序列准确无误,最终获得表达载体pET28a-AF0699。
1.3 重组蛋白的诱导表达和纯化 将AfuRecJ重组表达载体pET28a-AF0699转入大肠杆菌Rosetta (DE3)感受态细胞中,挑取单克隆于LB液体培养基(含50 μg/mL卡那霉素)中, 并将菌体逐步扩大培养至250 mL。待OD600达到0.6–0.8时,加入终浓度0.5 mmol/L的IPTG (isopropy-β-D-thiogalactoside,异丙基硫代半乳糖苷),20 ℃培养16 h,诱导重组蛋白表达。8000 r/min离心4 min收集菌体。将菌体沉淀悬浮30 mL裂解缓冲液(20 mmol/L Tris-HCl pH 8.0,300 mmol/L NaCl,10%甘油,5.0 mmol/L β-巯基乙醇,1.0 mmol/L PMSF),置于冰上超声破碎法裂解菌体:600 W功率下超声3 s,间歇3 s,超声15 min。在4 ℃下、10000 r/min离心30 min,收集细胞裂解上清液。预先用5倍树脂体积的裂解缓冲液在4 ℃下平衡Ni-NTA树脂1 h。将上清倒入已平衡好的树脂中,分别用含有5、10、20、50 mmol/L咪唑的裂解缓冲液梯度洗涤树脂,除去非特异性结合的杂蛋白,再用5 mL洗脱缓冲液(20 mmol/L Tris-HCl pH 8.0,300 mmol/L NaCl,300 mmol/L咪唑,10%甘油,5.0 mmol/L β-巯基乙醇,1.0 mmol/L PMSF)洗涤树脂,分步收集目标蛋白(1 mL/管)。通过SDS-PAGE检测纯度后,利用超滤管离心交换缓冲液,最终置于储存缓冲液(20 mmol/L Tris-HCl pH 7.5,100 mmol/L NaCl,50%甘油)中,于–20 ℃保存。蛋白浓度经Bradford法测定。
1.4 AfuRecJ核酸酶活性鉴定 用于检测AfuRecJ核酸酶活性的寡核苷酸片段见表 1,于–20 ℃避光储存。AfuRecJ核酸酶活性测定反应体系(10 μL)包括:20 mmol/L Tris-HCl pH 7.5,50 mmol/L NaCl,2.0 mmol/L MnCl2,1.0 mmol/L DTT,100 ng/μL BSA,100 nmol/L底物和1.67 μmol/L的AfuRecJ核酸酶。55 ℃反应20 min。加入10 μL反应终止液(95%甲酰胺,100 mmol/L EDTA,0.2% SDS,0.02%溴酚蓝)。反应产物于含有8 mol/L尿素的15%变性聚丙烯酰胺凝胶中电泳,经Amersham Typhoon RGB激光成像仪(GE公司)扫描成像并分析。
2 结果和分析 2.1 AfuRecJ核酸酶表达纯化、活性鉴定 将表达质粒pET28a-AF0699在E. coli Rosetta (DE3)中重组表达AfuRecJ蛋白,纯化结果见图 1-A。20 ℃诱导过夜,经镍柱亲和层析纯化,从250 mL培养液中得到约4 mg的AfuRecJ蛋白。经15% SDS-PAGE初步检测表明目的蛋白纯度达90%以上。通过与蛋白分子量标准对比,AfuRecJ蛋白的单体分子量约为50.0 kDa,与理论计算值(49.589 kDa)相符(图 1-A)。对其进行体外酶学活性的检测实验,在反应体系中加入梯度稀释的AfuRecJ,55 ℃反应20 min,结果表明AfuRecJ具有单链DNA核酸酶活性(图 1-B)。
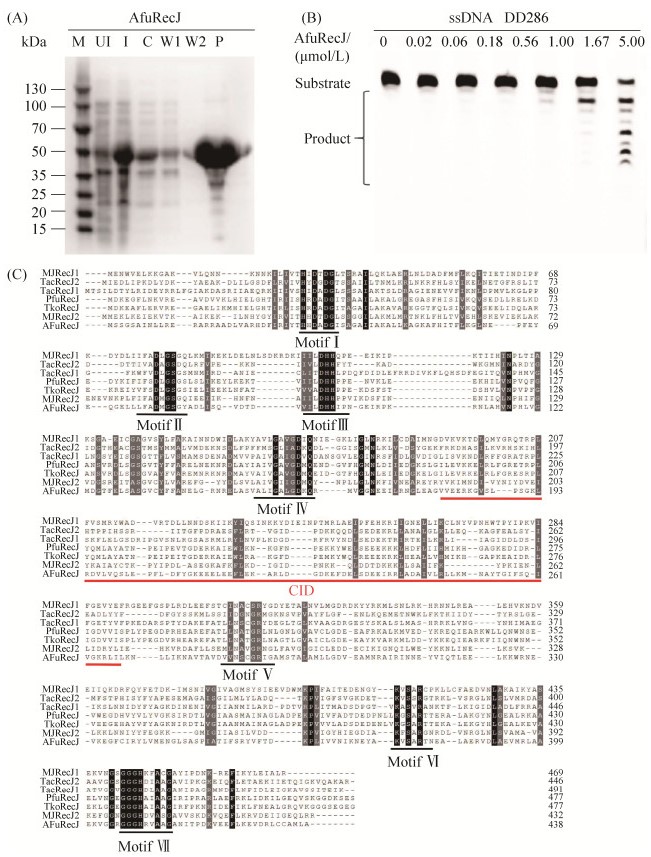 |
| 图 1 AfuRecJ表达纯化及活性测定 Figure 1 Expression and purification of AfuRecJ and activity titration. A: 15% SDS-PAGE analysis of recombinant AfuRecJ recovered from induced E. coli cells. The gel was stained with Coomassie blue R-250. Lane M: molecular weight marker; lane I and UI: induced and uninduced E. coli total proteins; lane C: supernatant after centrifugation; lane W1 and W2: wash through buffer B with 5.0 mmol/L and 50 mmol/L of imidazole; lane P: purified recombinant AfuRecJ. B: activity titration of AfuRecJ. The reaction mixtures contained 100 nmol/L ssDNA substrate, and increasing AfuRecJ in reaction buffer consisted of 20 mmol/L Tris-HCl pH 7.5, 50 mmol/L NaCl, 2.0 mmol/L MnCl2, 1.0 mmol/L DTT and 100 ng/μL BSA. The reactions were performed at 55 ℃ for 20 min. C: multi-alignment of different archaeal RecJs. The conserved motifs are marked by black lines, and the domain of CID (CMG Interaction Domain) is highlighted with the red line. |
| 图选项 |
2.2 AfuRecJ核酸酶活性测定条件的优化 初步鉴定AfuRecJ具有核酸酶活性,进一步对其反应条件进行优化,结果如图 2所示。AfuRecJ在pH 7.5的Tris-HCl缓冲体系中活性最高(图 2-A)。AfuRecJ酶促反应需要二价金属离子的参与,Mn2+和Mg2+均可促进AfuRecJ的核酸酶活性,其中Mn2+促进效果更为明显,其最优浓度是0.5 mmol/L (图 2-B)。图 2-C表明,AfuRecJ在低盐浓度下具有更高的活性,最适反应浓度为10 mmol/L,当NaCl浓度高于200 mmol/L时,AfuRecJ的核酸酶活性显著降低。AfuRecJ在65 ℃活性最高,在温度高于85 ℃后基本失活(图 2-D)。
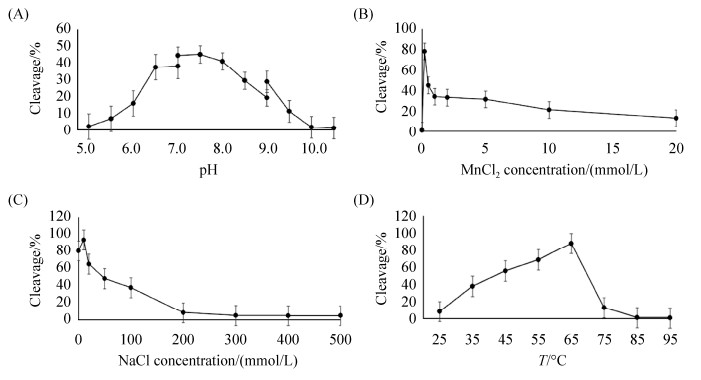 |
| 图 2 AfuRecJ水解单链DNA反应条件优化 Figure 2 Optimization of ssDNA hydrolysis by AfuRecJ. pH value (A), concentration of divalent manganese ion (B), ion strength (C), and reaction temperature (D) were optimized for the nuclease activity of AfuRecJ using a 23 nt single-stranded DNA (ssDNA) as substrate (100 nmol/L). The degraded amount of substrate DNA was quantified and plotted vs. each effect factor. |
| 图选项 |
2.3 AfuRecJ核酸酶的底物特异性 细菌的RecJ核酸酶仅从5?端水解单链DNA,而部分古菌RecJ同源蛋白能同时水解DNA和RNA[15–17],故分别利用单链DNA和RNA作为底物,检测AfuRecJ的核糖选择性。结果显示,AfuRecJ可以水解单链的DNA和RNA,相同条件下对RNA的水解活性更强(图 3)。
 |
| 图 3 AfuRecJ核酸酶的核糖选择性 Figure 3 Preferences for oligoribonucleotides of AfuRecJ nuclease. Reactions were performed at 55 ℃ by incubating increasing concentration of AfuRecJ with 100 nmol/L 5′-FAM-labeled 23 nt ssDNA (A) or ssRNA (B), respectively. |
| 图选项 |
2.4 AfuRecJ核酸酶的水解极性 使用全硫代修饰的单链DNA和RNA作为底物,检测AfuRecJ核酸酶的水解极性。图 4-A两种DNA底物长度相同,荧光标记在不同末端(DD305荧光标记在5?末端,DD306荧光标记位于3?末端)。AfuRecJ降解5?末端带有标记的单链DNA时产生梯度长度的寡核苷酸,在降解3?末端标记的DNA时只产生1 nt的产物。该结果表明,AfuRecJ对于单链DNA的降解是从3?末端起始的。图 4-B中RNA底物的荧光基团同样是标记在单链RNA的不同末端,AfuRecJ对于两种标记的单链RNA都能降解产生梯度长度的产物片段,表明AfuRecJ可从两端降解单链RNA,且3?–5?外切酶活性明显高于5?–3?外切酶活性。因此,对于单链DNA底物,AfuRecJ是3?外切酶;对于单链RNA底物,AfuRecJ主要表现为3?外切酶,同时具有微弱的5?外切酶活性。
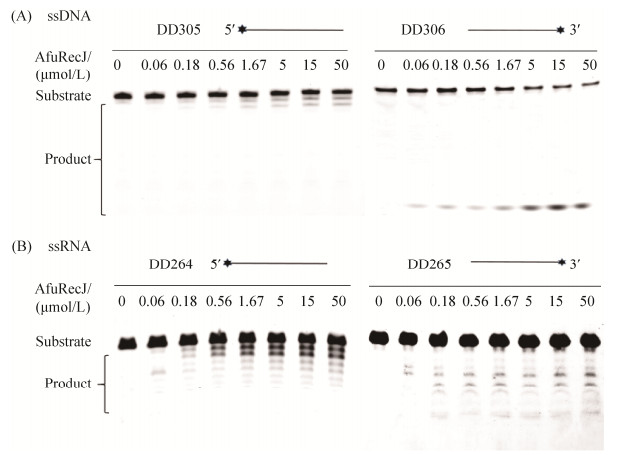 |
| 图 4 AfuRecJ核酸酶的水解极性 Figure 4 Hydrolysis polarity of oligonucleotides and oligodeoxynucleotide by AfuRecJ nuclease. AfuRecJ was incubated with 100 nmol/L fully phosphothioate-modified 23 nt ssDNA (A) or 17 nt ssRNA (B) labeled with a fluorescence group FAM at 5?- or 3?-end, respectively. |
| 图选项 |
2.5 底物末端基团磷酸化对AfuRecJ核酸酶活性的影响 使用3?末端磷酸化修饰的单链DNA和RNA作为底物,检测其对AfuRecJ核酸酶水解活性的影响。对于单链DNA底物,底物3?末端磷酸化对其活性有微弱的抑制作用(图 5-A)。对于单链RNA底物,底物3?末端的磷酸化对其核酸酶活性的抑制作用较为明显(图 5-B)。
 |
| 图 5 3?末端磷酸化对AfuRecJ核酸酶活性的影响 Figure 5 The effect of the 3?-terminal phosphate group on the AfuRecJ nuclease activity. AfuRecJ was incubated with 100 nmol/L 5?-FAM-labeled 23 nt ssDNA (A) or 12 nt ssRNA (B) which has a phosphate or hydroxyl group at 3?-end, respectively. |
| 图选项 |
2.6 AfuRecJ核酸酶水解底物长度的偏好性 AfuRecJ对不同长度的底物水解活性也不同。对于单链DNA底物而言,AfuRecJ不能有效水解长度小于等于6 nt的底物,对于长度为12–42 nt的底物水解活性较强,对于长度大于42 nt的底物水解活性略有降低(图 6-A)。对于单链RNA底物,AfuRecJ水解活性总体上强于DNA底物,对长度为4–16 nt的RNA底物水解活性都较强(图 6-B)。
 |
| 图 6 AfuRecJ核酸酶对底物长度的偏好性 Figure 6 Substrate length preferences of AfuRecJ. The AfuRecJ (1.67 μmol/L) was incubated with 5?-FAM-labeled ssDNA (A) or ssRNA (B) with different lengths at 55 ℃ for 20 min. |
| 图选项 |
2.7 AfuRecJ核酸酶对不同结构的DNA链的选择性 最后检测了AfuRecJ对不同二级结构的DNA底物的水解活性(图 7)。总体上,AfuRecJ水解单链DNA的活性最强。对于3?端特殊结构的双链DNA底物,水解活性从强到弱的顺序为3?突出(3?overhang) > 3?叉状(3?fork) > > 3?平端(3?blunt)=3?凹陷(3?recess),且对于3?平端(3?blunt)和凹陷(3?recess)结构基本无活性。
 |
| 图 7 AfuRecJ核酸酶对不同二级结构的DNA链的选择性 Figure 7 Selectivity of the AfuRecJ on DNA secondary structure. DNA with different secondary structures (single-stranded, forked, overhanged, recessed and blunt) were incubated with 5 μmol/L AfuRecJ at 55 ℃ for 20 min. |
| 图选项 |
3 讨论 现有研究表明细菌RecJ核酸酶仅具有5?–3?的单链DNA特异性外切酶,主要参与DNA同源重组和错配修复[4, 18]。生物信息学分析结果表明广古菌、奇古菌、初古菌都编码RecJ核酸酶同源蛋白,但泉古菌中没有发现RecJ同源蛋白。同时古菌编码RecJ核酸酶的数量和酶活性存在显著多样化。部分古菌仅存在一个RecJ蛋白,如嗜热球菌P. furiosus RecJ和T. kodarensis GAN。PfuRecJ具有单链DNA底物5?–3?外切酶、单链RNA底物3?–5?外切酶两种活性[15];而TkoGAN仅检测到单链DNA底物5?–3?外切酶活性,未检测到对RNA底物的切割活性[16]。另外部分古菌则含有多个RecJ核酸酶。甲烷球菌M. jannaschii较长的MjRecJ1具有5?–3?的外切核酸酶活性,较短的MjRecJ2主要表现为3?–5?的外切酶活性[17]。Thermoplasma acidophilum (T. acidophilum)较长的TacRecJ1具有ssDNA特异性的5?–3?外切酶活性;较短的TacRecJ2对ssDNA与ssRNA均为3?–5?的外切酶活性,且偏好ssRNA底物[19]。
与这些已鉴定的古菌RecJ核酸酶的酶学特征相比,AfuRecJ与TacRecJ2更为相似。二者都具有依赖于Mn2+的ssDNA和ssRNA依赖性3?–5?外切核酸酶活性,可水解单链DNA和RNA,对RNA的水解活性强于DNA。虽然AfuRecJ也能够从单链RNA的5?端微弱地水解核苷酸,但该活性结果可能是由于本文所用单链RNA底物的3?端的荧光标记阻碍了其从3?端降解,从而使其从5?端降解单链RNA。同时后续研究会继续尝试AF_0735在大肠杆菌重组表达,获得高浓度可溶性蛋白后,增高反应体系中蛋白浓度,鉴定其是否具有对ssDNA与ssRNA的水解活性。
细菌RecJ的催化活性主要由N端的催化核心(DHH和DHHA1两个结构域)[9]、C端的寡核苷酸/寡糖折叠结构域(OB)组成。部分细菌RecJ在C端还具有一个功能未知的多样化结构域[5, 11]。与细菌的RecJ核酸酶相比,古菌RecJ核酸酶在DHH结构域的基序IV和V之间(图 1-C)存在一个类似真核Cdc45蛋白所具有的CID结构域(CMG interaction domain,CMG相互作用结构域)[20]。真核Cdc45蛋白的CID结构域在真核细胞中参与MCM的结合,与GINS蛋白一同形成Cdc45-MCM-GINS (CMG)复合物,该复合物作为复制型解螺旋酶,对于DNA复制的起始起重要作用[20]。在古菌PfuRecJ和TkoGAN三级结构的DHH和DHHA1结构域之间也存在一个独立CID结构域[21]。与真核Cdc45蛋白不同的是,PfuRecJ和TkoGAN是通过DHH结构域与GINS蛋白相互作用,进而形成CMG复合物,其CID结构域的敲除并不影响二者的RecJ/GAN与GINS形成复合物。古菌M. jannaschii的两个RecJ核酸酶也具有CID结构域,然而二者均不能和GINS蛋白形成复合物。而同样具有CID结构域的T. acidophilum的2个RecJ核酸酶中,只有TacRecJ2可以与GINS形成复合物。另外泉古菌门的硫化叶菌含有和真核生物Cdc45类似的蛋白(古菌Cdc45)。硫化叶菌Cdc45、MCM、GINS形成类似真核生物的CMG复合物,在DNA复制中行使DNA解螺旋酶功能[22]。古菌的RecJ蛋白普遍具有核酸酶活性,而真核和古菌的Cdc45蛋白则不具有核酸酶活性[20, 22]。虽然AfuRecJ核酸酶中也存在类似的CID结构域(图 1-C),但生信序列分析并未发现A. fulgidus中有GINS蛋白,导致无法鉴定AfuRecJ与GINS是否形成复合物。我们将在后续研究中寻找A. fulgidus GINS蛋白的替代蛋白,研究其与AfuRecJ的相互作用。
本研究阐明了AfuRecJ在体外的生化功能和酶学特征,对于其体内功能的鉴定还需要进一步的实验证明。后期将从其晶体结构角度解析优先水解单链RNA的催化机制,以及鉴定GINS蛋白的功能同源蛋白,并验证其是否参与形成MCM-GINS-RecJ复合物及其功能。鉴于古菌RecJ通常能够同时水解DNA和RNA,可能参与RNA代谢,因此对于A. fulgidus RecJ核酸酶的研究有利于深入理解古菌的DNA复制与损伤修复以及RNA加工成熟与降解循环机制。
References
| [1] | Yang W. Nucleases:diversity of structure, function and mechanism. Quarterly Reviews of Biophysics, 2011, 44(1): 1-93. |
| [2] | Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Current Biology, 1994, 4(12): 1069-1076. DOI:10.1016/S0960-9822(00)00245-1 |
| [3] | Makarova KS, Koonin EV, Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biology Direct, 2012, 7(1): 7. DOI:10.1186/1745-6150-7-7 |
| [4] | Thoms B, Borchers I, Wackernagel W. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. Journal of Bacteriology, 2008, 190(1): 179-192. DOI:10.1128/JB.01052-07 |
| [5] | Wakamatsu T, Kitamura Y, Kotera Y, Nakagawa N, Kuramitsu S, Masui R. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. Journal of Biological Chemistry, 2010, 285(13): 9762-9769. DOI:10.1074/jbc.M109.096487 |
| [6] | Sutera VA Jr, Han ES, Rajman LA, Lovett ST. Mutational analysis of the RecJ exonuclease of Escherichia coli:identification of phosphoesterase motifs. Journal of Bacteriology, 1999, 181(19): 6098-6102. DOI:10.1128/JB.181.19.6098-6102.1999 |
| [7] | Yamagata A, Masui R, Kakuta Y, Kuramitsu S, Fukuyama K. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Research, 2001, 29(22): 4617-4624. DOI:10.1093/nar/29.22.4617 |
| [8] | Rajman LA, Lovett ST. A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. Journal of Bacteriology, 2000, 182(3): 607-612. DOI:10.1128/JB.182.3.607-612.2000 |
| [9] | Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(9): 5908-5912. DOI:10.1073/pnas.092547099 |
| [10] | Pellegrini L. Structural insights into Cdc45 function:was there a nuclease at the heart of the ancestral replisome?. Biophysical Chemistry, 2017, 225: 10-14. DOI:10.1016/j.bpc.2016.11.011 |
| [11] | Cheng KY, Xu H, Chen XY, Wang LY, Tian B, Zhao Y, Hua YJ. Structural basis for DNA 5'-end resection by RecJ. eLife, 2016, 5: e14294. DOI:10.7554/eLife.14294 |
| [12] | Li MJ, Yi GS, Yu F, Zhou H, Chen JN, Xu CY, Wang FP, Xiao X, He JH, Liu XP. The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45. Nucleic Acids Research, 2017, 45(21): 12551-12564. DOI:10.1093/nar/gkx887 |
| [13] | Oyama T, Ishino S, Shirai T, Yamagami T, Nagata M, Ogino H, Kusunoki M, Ishino Y. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Research, 2016, 44(19): 9505-9517. DOI:10.1093/nar/gkw789 |
| [14] | Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, Richardson DL, Kerlavage AR, Graham DE, Kyrpides NC, Fleischmann RD, Quackenbush J, Lee NH, Sutton GG, Gill S, Kirkness EF, Dougherty BA, McKenney K, Adams MD, Loftus B, Peterson S, Reich CI, McNeil LK, Badger JH, Glodek A, Zhou LX, Overbeek R, Gocayne JD, Weidman JF, McDonald L, Utterback T, Cotton MD, Spriggs T, Artiach P, Kaine BP, Sykes SM, Sadow PW, D'Andrea KP, Bowman C, Fujii C, Garland SA, Mason TM, Olsen GJ, Fraser CM, Smith HO, Woese CR, Venter JC. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature, 1997, 390(6658): 364-370. DOI:10.1038/37052 |
| [15] | Yuan H, Liu XP, Han Z, Allers T, Hou JL, Liu JH. RecJ-like protein from Pyrococcus furiosus has 3'-5' exonuclease activity on RNA:implications for proofreading of 3'-mismatched RNA primers in DNA replication. Nucleic Acids Research, 2013, 41(11): 5817-5826. DOI:10.1093/nar/gkt275 |
| [16] | Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Research, 2011, 39(14): 6114-6123. DOI:10.1093/nar/gkr181 |
| [17] | Yi GS, Song Y, Wang WW, Chen JN, Deng W, Cao WG, Wang FP, Xiao X, Liu XP. Two archaeal RecJ nucleases from Methanocaldococcus jannaschii show reverse hydrolysis polarity:implication to their unique function in Archaea. Genes, 2017, 8(9): 211. DOI:10.3390/genes8090211 |
| [18] | Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(12): 6765-6770. DOI:10.1073/pnas.121183298 |
| [19] | Ogino H, Ishino S, Kohda D, Ishino Y. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3?-5? exonuclease that associates with a DNA replication complex. Journal of Biological Chemistry, 2017, 292(19): 7921-7931. DOI:10.1074/jbc.M116.767921 |
| [20] | Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nature Communications, 2016, 7(1): 11638. |
| [21] | Nagata M, Ishino S, Yamagami T, Ogino H, Simons JR, Kanai T, Atomi H, Ishino Y. The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis. Nucleic Acids Research, 2017, 45(18): 10693-10705. DOI:10.1093/nar/gkx740 |
| [22] | Xu YL, Gristwood T, Hodgson B, Trinidad JC, Albers SV, Bell SD. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(47): 13390-13395. DOI:10.1073/pnas.1613825113 |
