王晓晓1#, 孙世博1#, 张意慈1, 张楠1,2, 陈继红1, 徐卫平3, 马强2, 许建强1


1. 大连理工大学生命科学与药学学院, 盘锦产业技术研究院, 辽宁 盘锦 124221;
2. 中国检验检疫科学研究院, 北京 100176;
3. 大连理工大学海洋科学与技术学院, 工业生态与环境工程教育部重点实验室, 辽宁 盘锦 124221
收稿日期:2019-10-20;修回日期:2019-11-17;网络出版日期:2019-11-26
基金项目:国家自然科学基金(31670767);中央高校基本科研业务费(DUT17JC36,DUT17RC(4)27);大连理工大学盘锦产业技术研究院研发项目(PJYJY-002-862011)
*通信作者:许建强, Tel:+86-427-2631418;E-mail:Jianqiang.Xu@dlut.edu.cn.
#共同第一作者。
摘要:[目的] 基于大肠杆菌掺硒机器,寻找tRNASec骨架上关键核苷酸位点,为解决硒蛋白目前面临的掺硒效率较低、产量低的问题提供新思路。[方法] 以大鼠细胞质型硫氧还蛋白还原酶(thioredoxin reductase 1,TrxR1)为掺硒模式蛋白为定点突变tRNASec,转化至BL21(DE3)gor-获得阳性重组菌株(携带pET-TRSter/pSUABC'),用于表达大鼠硒蛋白TrxR1,然后使用2',5'ADP-Sepharose亲和层析和凝胶过滤两步法分离纯化TrxR1,最后利用经典硒依赖型DTNB还原反应测定TrxR1的酶活,分析关键核苷酸位点,评价掺硒效率。[结果] 在存在SECIS元件的前提下,当SelA、SelB、tRNASec共表达时,与野生型相比,携带突变型tRNASec所共表达的TrxR1酶活力呈现不同程度的降低,其中E.coli tRNASec的G18、G19这两个位点的所有的TrxR1酶活远低于野生型(< 10%);然而,a26和b7的酶活相对较高。[结论] E.coli tRNASec骨架上G18和G19位点对于维持tRNA稳定性和灵活性发挥了关键作用,位点突变引起tRNA结构变化会影响tRNASec与掺硒元件的互作,因此有望通过改造tRNA核苷酸位点来提高硒蛋白的掺硒效率。
关键词:大肠杆菌硒代半胱氨酸硒蛋白硒代半胱氨酸特异性tRNASec细胞质型硫氧还蛋白还原酶
Key nucleotide sites of Escherichia coli tRNASec
Xiaoxiao Wang1#, Shibo Sun1#, Yici Zhang1, Nan Zhang1,2, Jihong Chen1, Weiping Xu3, Qiang Ma2, Jianqiang Xu1


1. School of Life and Pharmaceutical Sciences(LPS), Panjin Institute of Industrial Technology, Dalian University of Technology, Panjin 124221, Liaoning Province, China;
2. Chinese Academy of Inspection and Quarantine, Beijing 100176, China;
3. School of Ocean Science and Technology(OST) and Key Laboratory of Industrial Ecology and Environmental Engineering(MOE), Dalian University of Technology, Panjin 124221, Liaoning Province, China
Received: 20 October 2019; Revised: 17 November 2019; Published online: 26 November 2019
*Corresponding author: Jianqiang Xu, Tel:+86-427-2631418;E-mail:Jianqiang.Xu@dlut.edu.cn.
Foundation item: Supported by the National Natural Science Foundation of China (31670767), by the Fundamental Research Funds for the Central Universities (DUT17JC36, DUT17RC(4)27) and by the Research and Development Program of Panjin Institute of Industrial Technology (DUT) (PJYJY-002-862011)
# These authors contributed equally to this work.
Abstract: [Objective] Based on the Sec insertion machinery in Escherichia coli, we aimed at finding key sites of tRNASec scaffold and providing a new way to solve the problems of low Sec insertion efficiency and low yield of selenoprotein. [Methods] We used rat cytoplasmic thioredoxin reductase (thioredoxin reductase 1, TrxR1) as a model selenoprotein. First, we constructed tRNASec by site-directed mutagenesis, and transformed them to BL21 (DE3) gor-to obtain a positive recombinant strain (carrying pET-TRSter'/pSUABC), which was used to express rat cytoplasmic thioredoxin reductase 1 (TrxR1). Then TrxR1 was purified using 2', 5'-ADP Sepharose affinity chromatography and gel filtration. The enzyme activities of TrxR1 were determined by the classical Se-dependent DTNB reduction assay to analyze the key nucleotide sites and evaluate the Sec incorporation efficiency. [Results] When tRNASec co-expressed with SelA and SelB in the presence of the bacterial SECIS element, the activities of TrxR1 decreased at varying degrees compared with wild type. Among them, the enzyme activities of all G18 and G19 mutants were much lower than that of wild type (< 10%). However, the enzymatic activities of a26 and b7 were relatively high. [Conclusion] G18 and G19 nucleotide sites of E. coli tRNASec may play a vital role in maintaining the stability and flexibility of tRNASec. Site-directed mutations of tRNASec causing the conformational change may affect the interaction between tRNASec and Sec elements. Thus, it is possible to improve the Sec insertion efficiency via proper modification on tRNA nucleotide sites.
Keywords: Escherichia coliselenocysteine (Sec)selenoproteinSec incorporation tRNASeccytoplasmic thioredoxin reductase (TrxR1)
硒蛋白(selenoprotein)广泛分布于自然界生物体中,参与调节机体氧化应激、细胞凋亡、维持细胞生长与细胞增殖等[1-2]。硒蛋白所含的硒代半胱氨酸(selenocysteine,Sec)是第21种氨基酸,由终止密码子UGA编码,具有低pKa值、高亲核性和高极性的性质,通常作为硒酶(selenoenzyme)的活性中心[3-4]。原核生物体内不存在硒代半胱氨酸合成酶,而是沿着“丝氨酸途径”进行硒蛋白的合成[5]。如图 1所示,首先SerRS催化tRNASec生成Ser-tRNASec,然后SelA催化Ser-tRNASec形成Sec-tRNASec,随后SelB识别并结合Sec-tRNASec和硒代半胱氨酸插入序列(selenocysteine insertion sequence,SECIS)元件,最终将Sec掺入到多肽链中[6]。
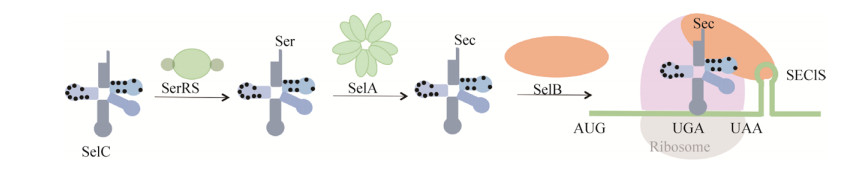 |
| 图 1 原核生物掺硒机制示意图 Figure 1 Schematic diagram of the Sec insertion mechanism in prokaryotes. Sec-specific tRNASec interacts with different factors such as SerRS, SelA and SelB accordingly. In the presence of bacterial SECIS element, tRNASec ultimately carries Sec to the small subunit of ribosome to incorporate Sec into growing polypeptides and eventually forms selenoproteins. In this study, we aimed to search for key nucleotide sites on E. coli tRNASec scaffold. |
| 图选项 |
然而,全长硒蛋白生物合成受到多因素限制。首先,释放因子RF2竞争性地影响SelB与UGA的识别与结合,限制了掺硒效率。其次,SerRS影响SelA与tRNASec结合效率,也会影响硒蛋白的品质[7]。再次,硒蛋白合成需要一个紧跟UGA、位于3'-UTR的SECIS掺硒元件,其与SelB可以发生特异性结合[8]。此外,不依赖SECIS元件合成硒蛋白成为一些****关注和研究的一个方向。Aldag等[9]构建的tRNAUTu突变体能够识别EF-Tu,减少了Sec掺入限制,但Ser等氨基酸的掺杂比例增加(35%–45%)[10]。后来,人们又构建了tRNAUTuX和tRNAUTuT6突变体,掺硒效率并未获得较大提升,但是含硒蛋白产率却降低了[11]。掺硒效率低和含硒蛋白产量低仍旧是当前研究难题,我们亟需寻找到硒蛋白高效生物合成的新方法和新途径。
tRNASec具有独特的二级结构,与多种元件相互作用,是掺硒机制中的核心元件[12-13]。与常规tRNA受体臂+TΨC臂“7+5”结构不同,细菌tRNASec为保守的“8+5”二级结构。常规tRNA的D臂由3–4 bp (base pair, bp)的D茎和7–12 nt (nucleotide,nt)的D环组成,而细菌tRNASec D臂是由6 bp的D茎和4 nt的D环构成[14]。Aquifex aeolicus SelA与Thermoanaerobacter tengcongensis tRNASec复合体晶体结构显示,SelA与tRNASec的D臂和受体-TΨC环结合[15]。Aquifex aeolicus SelB与Sec-tRNASec的复合体晶体结构显示,SelB与tRNASec的T臂结合[16]。即沿着丝氨酸途径合成硒蛋白将阻止tRNASec与EF-Tu结合,减少Ser掺入到Sec中,促进原核生物产生高品质的硒蛋白。因此,我们可以从改造tRNA的D臂或T臂入手,找出tRNA关键核苷酸位点。荧光各向异性结合测定结果表明,E. coli tRNASec能够与E. coli SelA、SelB结合[16-17]。由于缺乏E. coli tRNASec与SelA (或SelB)的完整的晶体结构,tRNASec与它们之间具体的核苷酸结合位点并不清楚,这将给寻找掺硒效率高的硒蛋白带来困难。
为了理解tRNASec与SelA、SelB间的相互作用及分子机制,在本研究中,我们利用定点突变技术构建tRNASec突变体。首先筛选出可能的位置,包括tRNASec的D臂和T臂;其次,选取E. coli tRNASec D臂上的G18、U20、A21以及T臂上的U63、G64、U65核苷酸位点进行多点突变,并对G18和G19位点进行单点突变;然后以大鼠硒蛋白TrxR1为模型,通过DTNB反应检测TrxR1活性[18]。最后对酶活相对较高的突变体进行纯化,验证TrxR1酶活稳定性。TrxR1催化是一种高度硒依赖型反应,通过评估TrxR1活性,反映掺硒效率。我们初步探究了维持E. coli tRNASec整体结构的关键核苷酸位点,这将完善大肠杆菌掺硒系统,为合成高活力硒蛋白提供有益参考,进一步推动了哺乳动物硒蛋白在癌症治疗等方面的研究[19-20]。
1 材料和方法 1.1 实验材料与仪器
1.1.1 菌株与质粒: 大肠杆菌DH5α菌株来自本实验室,大肠杆菌BL21 (DE3) gor-(基因组上已敲除谷胱甘肽还原酶基因,四环素抗性)由瑞典卡罗林斯卡医学院Arne Holmgren教授馈赠,pCDF-SelC (携带E. coli selC基因,链霉素抗性)质粒、pET-TRSter (携带大鼠野生型TrxR1基因,卡那抗性)质粒、pSUABC (携带selA、selB和selC基因,氯霉素抗性)均由瑞典卡罗林斯卡医学院Elias Arnér教授馈赠,由本实验室保藏并开展研究。
1.1.2 主要试剂: 基因操作试剂盒如Gene JET Gel Extraction Kit、Gene JET PCR Purification Kit、Gene JET Miniprep Kit、限制性内切酶(包括Restriction Enzyme Eco31 I、Dpn I、Acc65 I、Xba I)以及T4连接酶均购自Thermo Fisher Scientific公司。DNA Marker购自宝生物工程(大连)有限公司(大连TaKaRa公司)。Bradford法蛋白浓度测定试剂盒购自生工生物工程(上海)股份有限公司。酶活底物5, 5'-Dithiobis-(2-nitrobenzoic acid) (DTNB)购自Sigma-Aldrich公司(USA)。
1.1.3 主要仪器: Biometra三单元及96孔Professional Thermocycler PCR仪购自德国Analytik JENA公司。全功能酶标仪SYNERGY H1购自美国BioTek公司。紫外分光光度计UV5 BIO购自瑞士Mettler Toledo公司。?KTA Start workstation、2', 5' ADP-SepharoseTM 4B和HiPrep 16/60 SephacrylTM S-300 High Resolution购自GE公司生命科学部门(GE Healthcare Life Sciences,Uppsala,Sweden)。Amicon?Ultra-15mL超滤离心管(30 kDa)购自Merck-Millipore公司(USA)。
1.2 构建tRNASec突变体和pSUABC'表达质粒 根据pCDF-SelC基因序列设计引物(表 1),在selC基因上引入定点突变[21-22]。以pCDF-SelC质粒为模板,进行反向PCR,用Dpn I/Eco31 I双酶切PCR产物,采用T4 DNA连接酶进行自连,获得重组质粒pCDF-SelC',DNA测序结果验证了在相应的位点上引入了预期突变。利用Acc65 I/ Xba I双酶切pCDF-SelC'以及pSUABC,分别切胶回收400 bp、7000 bp左右处片段,用T4 DNA ligase连接,获得pSUABC'重组表达质粒。引物合成和DNA序列测定委托生工生物工程(上海)股份有限公司完成。
表 1. 定点突变所用的引物 Table 1. Primers for site-directed mutagenesis
| Primer | Sequence (5'→3')1 | Purpose2 |
| F05A | GCGGTCTCGTCTCCAGTGAGGCGGCTGGACTTC | Forward primer for g18a mutant |
| F05U | GCGGTCTCGTCTCCTGTGAGGCGGCTGGACTTC | Forward primer for g18u mutant |
| F05C | GCGGTCTCGTCTCCCGTGAGGCGGCTGGACTTC | Forward primer for g18c mutant |
| F07A | GCGGTCTCGTCTCCGATGAGGCGGCTGGACTTC | Forward primer for g19a mutant |
| F07U | GCGGTCTCGTCTCCGTTGAGGCGGCTGGACTTC | Forward primer for g19u mutant |
| F07C | GCGGTCTCGTCTCCGCTGAGGCGGCTGGACTTC | Forward primer for g19c mutant |
| F17 | GCGGTCTCGTCTCCNGNGNGGCGGCTGGACTTC | Forward primer for rma mutants |
| F18 | GCGGTCTCGACTCCNNNGATCTTCCGCCAAAATGC | Forward primer for rmb mutants |
| R05 | GCGGTCTCGGAGACGACGATCTTCCGCGCC | Reverse primer for g18, g19, and rma mutants |
| R08 | GCGGTCTCGGAGTCGAACCTGCCCGGGAC | Reverse primer for rmb mutants |
| T7 | TAATACGACTCACTATAGGG | Forward primer for sequencing |
| T7TER | TGCTAGTTATTGCTCAGCGG | Reverse primer for sequencing |
| This table lists the primers used for introducing mutations at the D arm or T arm of E. coli tRNASec (selC gene product) scaffold, using the wild type pCDF-SelC plasmid as template. 1 Restriction cleavage sites (Eco31 I) in primer sequences were underlined. N is a degenerate base (A/C/G/U). 2 RMA were mutants constructed by introducing degenerate base N at G18, U20 or A21 sites of E. coli tRNASec. And RMB indicates mutants constructed by introducing N at U63, G64 or U65 sites of E. coli tRNASec. | ||
表选项
1.3 TrxR1蛋白的表达与纯化 将pSUABC'质粒转化到BL21 (DE3) gor- (含pET-TRSter质粒)感受态中,涂布于TCK抗性固体LB平板上,平板含50 μg/mL卡那霉素(Kana,K)、34 μg/mL氯霉素(Chl, C)、15 μg/mL四环素(Tet,T)。37 ℃过夜培养,之后挑取生长状态良好的圆形单菌落至5 mL含TCK抗性的新鲜液体LB培养基中,于37 ℃、220 r/min振荡培养12 h。按1%的接种量将菌液接种到24孔板(含3 mL的TCK抗性LB液体培养基)、100 mL锥形瓶(含20 mL的TCK抗性LB液体培养基)或1 L锥形瓶(含200 mL的TCK抗性LB液体培养基),于37 ℃、220 r/min振荡培养至对数生长末期(OD600nm≈2.4)。依次加入0.5 mmol/L IPTG、5 μmol/L selenite、100 mg/L L-cysteine(勿要同时加入),于24 ℃、220 r/min继续振荡培养24 h。4000 r/min离心20 min,收集菌体,用TE buffer (pH 7.5) (50 mmol/L Tris-HCl+20 mmol/L EDTA)重悬,待用[23]。
1.4 TrxR1蛋白的分离纯化 向重悬液中加入溶菌酶(50 mg/mL)至终浓度为1 mg/mL,混匀,反复冻融3次后,超声破碎。4℃、13000 r/min离心20 min后,取上清,经0.45 μm无菌滤膜过滤后,上样至2', 5' ADP-Sepharose亲和层析柱纯化(利用24孔板表达的TrxR1不进行纯化)。纯化后的样品利用超滤管将其浓缩至3 mL,于4 ℃、13000 r/min离心20 min,取2 mL上清上样到凝胶过滤柱,用含有0.15 mol/L NaCl的50 mmol/L Tris-HCl (pH 7.5)平衡后进行连续线性梯度洗脱。蛋白凝胶过滤与?KTA StartTM蛋白纯化工作站联用,通过监测UV 280 nm (蛋白质吸光度)自动收集洗脱峰,每管收集4 mL,对自动收集的组分测定其在463 nm处吸光度值,TrxR1蛋白样品于4 ℃保存[24]。
1.5 TrxR1蛋白浓度的测定 利用Bradford法在96孔板测定蛋白浓度,每个标准品和样品设置3个孔平行测定,以牛血清蛋白作为标准蛋白,测定波长设置在595 nm。将500 μL TE buffer (空白)或纯酶放入石英比色皿中,利用紫外分光光度计测定OD463处的FAD吸光度值,以此来计算TrxR1纯酶浓度,其中,FAD的摩尔吸光系数为11300 M–1 cm–1。
1.6 SDS-PAGE 将蛋白样品与5×SDS Loading Buffer混合,后者含有250 mmol/L Tris-HCl (pH 6.8)、10% SDS (M/V)、5% β-巯基乙醇(V/V)、50%甘油(V/V)和0.5%溴酚蓝(M/V)。混合样品在100 ℃加热10 min,之后离心,上样,进行10%还原型SDS-PAGE凝胶电泳,凝胶用考马斯亮蓝R250染色显示蛋白条带,使用凝胶系统记录电泳结果。
1.7 TrxR1酶活力测试 在96孔酶标板中加入5 μL TrxR1,随后加入195 μL DTNB反应液,DTNB反应液包括2.5 mmol/L DTNB、300 μmol/L NADPH、50 mmol/L TE Buffer (pH 7.5)。使用BioTek酶标仪在412 nm下连续监测96孔酶标板中TNB-的生成量,选取反应初始阶段线性部分,根据线段斜率进行酶活力计算[6]。本文中,在25 ℃条件下,TrxR1催化还原DTNB,我们把每分钟消耗1 μmol/L TNB定义为1个酶活单位(U),体积活力表述为U/mL蛋白,比活力表述为U/mg蛋白。其中,TNB-的摩尔吸光系数为13600 mol/(L·cm)。所有酶促反应均在室温(25 ℃)条件下测试,读数间隔10,总时长3 min,以未加酶的反应液为空白对照,至少重复3次测量,使用GraphPad Prism 7.0软件进行分析。
1.8 掺硒效率测试的统计方法 本研究中显示的所有实验数据均为3–5个独立实验的平均值±标准偏差(mean±SD)。数据采用GraphPad Prism version 7.0软件(GraphPad软件,San Diego,USA)进行分析。
2 结果和分析 2.1 构建E. coli tRNASec突变体 为了确定tRNASec不同位点的突变对酶活的影响,我们进行tRNASec突变体的构建,突变位点详见表 2。首先,在selC基因的“G18、U20、A21”和“U63、G64、U65”位点处,分别使用简并引物引入碱基突变,得到了16个多点突变体。然后对大肠杆菌tRNASec的G18和G19位点进行其他3个碱基的单点突变,共产生G18A、G18U、G18C、G19A、G19U和G19C这6个单点突变体。最后,我们构建了22个pCDF-SelC’和pSUABC'重组质粒。
表 2. E. coli tRNASec多点突变 Table 2. Multipoint mutations of E. coli tRNASec
| Mutant | Mutation sites | Mutant | Mutation sites | |
| RMA1 | U20C; A21G | RMA26 | U20A; A21G | |
| RMA5 | G18U; A21U | RMA28 | G18A; A21G | |
| RMA6 | G18U; U20C | RMA29 | G18A; U20A; A21G | |
| RMA11 | C16G; G20A; ΔU20 | RMB1 | U63C; G64A | |
| RMA18 | G18C; U20A; A21C | RMB2 | U63C; G64A; U65C | |
| RMA22 | G18C; U20C; A21G | RMB7 | G64U; U65C | |
| RMA24 | G18U; U20C; A21G | RMB11 | U63G; U65G | |
| RMA25 | G18A; U20C | RMB12 | U63G; G64C; U65G | |
| Numbers denote the position of mutation on the Escherichia coli tRNASec scaffold. A, U, G, C represent for adenine, uracil, guanine, and cytosine. “Δ” means deletion. | ||||
表选项
2.2 E. coli tRNASec的核苷酸位点改变对TrxR1酶活的影响 原核生物tRNASec是掺硒系统的重要因子,与SelA和SelB分别发生相互作用,如图 1所示。我们推测改变大肠杆菌tRNASec可能会改变掺硒效率。为了分析这些tRNA突变体的特性,在SECIS元件存在的情况下,我们先利用22个selC突变株共表达TrxR1,也就是将pSUABC’转化到BL21 (DE3) gor-(含pET-TRSter质粒)感受态中,在24深孔板中表达TrxR1,然后通过DTNB反应检测TrxR1粗酶的体积活力(图 2)。从考马斯亮蓝染色分析可以看出,55 kDa处有明显的亚基条带,TrxR1获得表达。
 |
| 图 2 TrxR1蛋白表达、纯化与酶活检测 Figure 2 Technical schematics of TrxR1 production: protein expression, purification and enzyme activity detection. Firstly, the pSUABC' plasmids carrying the tRNASec mutants (selC' genes) were transformed into BL21 (DE)3 gor- (pET-TRSter) strain. TrxR1 were obtained and further analyzed by DTNB reduction assay using an enzyme-labeling microplate reader. Finally, the mutants with higher enzyme activities were identified by the data analysis. |
| 图选项 |
从图 3可以看出,突变体tRNA的均低于野生型tRNA共表达的TrxR1酶活。与其他突变体相比,a26和b7的酶活相对较高。结果表明,改变大肠杆菌tRNASec核苷酸位点能够调节TrxR1酶活,这将有利于获得潜在的高活性硒蛋白。而有关G18和G19位点的所有单点突变体,它们共表达的TrxR1酶活均低于野生型的10% (表 3)。tRNASec上的G18和G19位点的突变导致硒蛋白酶活丧失,可能是大肠杆菌tRNASec重要位点,关系到掺硒效率,可以进一步研究。
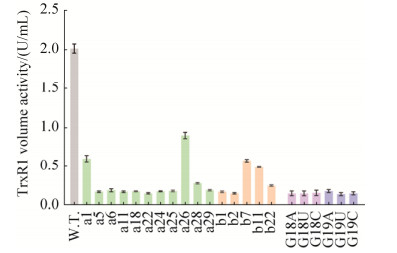 |
| 图 3 使用tRNASec突变体制备并纯化的TrxR1进行酶活分析(24深孔板) Figure 3 Enzyme activity analysis of TrxR1 produced with site-directed mutant tRNASec (cultured in the 24 deep-well plates). All the samples were induced by IPTG at a final concentration of 0.5 mmol/L and cultured in the 24 deep-well plates. The TrxR1 crude enzyme activities were measured by the DTNB assay, and the results are shown in Table 2. Data are expressed as mean±SD. W.T. means wild-type TrxR1. "a" represents carrying RMA tRNASec mutants co-expressing TrxR1. Similarly, "b" denotes carrying RMB tRNASec mutants co-expressing TrxR1. |
| 图选项 |
表 3. DTNB还原检测分析 Table 3. DTNB reduction assay
| Sample | Volume activity */(U/mL) | Sample | Volume activity */(U/mL) | Sample | Volume activity */(U/mL) | ||
| W.T. | 2.00 | W.T., -IPTG | 0.10 | a1 | 0.61 | ||
| a5 | 0.18 | a6 | 0.20 | a11 | 0.18 | ||
| a18 | 0.19 | a22 | 0.16 | a24 | 0.19 | ||
| a25 | 0.19 | a26 | 0.90 | a28 | 0.29 | ||
| a29 | 0.20 | b1 | 0.18 | b2 | 0.16 | ||
| b7 | 0.58 | b11 | 0.50 | b22 | 0.26 | ||
| G18A | 0.16 | G18U | 0.16 | G18C | 0.17 | ||
| G19A | 0.19 | G19U | 0.15 | G19C | 0.16 | ||
| This table summarized the enzyme activities (volume activities) of these TrxR1 produced with tRNASec variants cultured in 24 deep-well plates. We resuspended E. coli BL21 (DE)3 gor- (pSUABC'/pET-TRSter) wet cell pellet with 2 mL TE buffer (pH 7.5). All samples were induced by adding 0.5 mmol/L IPTG, except “W.T.-IPTG” which meant without IPTG. * TrxR1 crude enzyme activities were determined by the standard DTNB reduction assay. Each group of data was measured three times and the average value was shown in the table. | |||||||
表选项
2.3 确认G18和G19是大肠杆菌tRNASec的关键位点 为了排除小体系表达蛋白的不稳定对结果造成影响,我们先在100 mL摇瓶中表达a26和b7、G18和G19单点突变体,然后利用2', 5' ADP-Sepharose亲和层析纯化TrxR1蛋白,同时检测TrxR1纯化前后的酶活。考马斯亮蓝染色分析显示,纯化前(图 4-A)目标带明显,纯化后(图 4-B)几乎只剩下目标带。
 |
| 图 4 使用tRNASec突变体制备并纯化的TrxR1进行还原型SDS-PAGE分析 Figure 4 Reducing SDS-PAGE analysis of purified TrxR1 produced with tRNASec variants. Rat TrxR1 were expressed by BL21 (DE)3 gor- (pSUABC'/pET-TRSter) strain, which were cultured in 100 mL shaking flask and further purified by 2', 5' ADP-Sepharose affinity chromatography. The crude sample (A) and the purified protein (B) were run on 10 % SDS-PAGE gels. "M" stands for MW protein maker. |
| 图选项 |
与野生型tRNASec相比,基于突变体tRNASec的TrxR1经过表达、纯化,硒蛋白产物的酶活性呈现不同程度的降低,如图 5、表 4所示,其中G18和G19突变体的体积活力则更低,减少到不足野生型的30%。这些数据表明,改变G18或G19位点不利于产生高活性的硒蛋白。tRNA是一种柔性结构,某些位点的改变可能会改变tRNA的整体结构,导致tRNA减少和某些作用因子的相互作用,从而降低掺硒效率,影响硒蛋白的功能。
 |
| 图 5 使用tRNASec突变体制备的TrxR1纯酶进行DTNB分析 Figure 5 The DTNB activity assay of the pure enzyme of TrxR1 produced with mutant tRNASec. A: All the samples were induced by IPTG at a final concentration of 0.5 mmol/L. Those TrxR1 produced with tRNASec mutants were expressed by E. coli BL21 (DE3) gor- (pSUABC'/pET-TRSter) stain in 100 mL shaking flask and purified via 2', 5' ADP-Sepharose affinity chromatography. The pure enzyme activities of TrxR1 were tested by DTNB reduction assay, and the measured data are shown in Table 4. B: The secondary structure of E. coli tRNASec. |
| 图选项 |
表 4. DTNB分析 Table 4. DTNB reduction assay
| Samplea | Supernatant volume activity/(U/mL)b | Eluted enzyme activity/(U/mg)c | Samplea | Supernatant volume activity/(U/mL)b | Eluted enzyme activity/(U/mg)c | |
| W.T.-IPTG | 0.04 | - | b7 | 0.18 | 2.06 | |
| W.T. | 0.83 | 7.51 | a26 | 0.22 | 2.51 | |
| G18A | 0.06 | 0.51 | G19A | 0.05 | 0.49 | |
| G18U | 0.06 | 1.19 | G19U | 0.05 | 0.50 | |
| G18C | 0.07 | 0.34 | G19C | 0.10 | 0.51 | |
| Rat TrxR1 proteins were expressed in E. coli BL21 (DE)3 gor- (pSUABC’ / pET-TRSter) strain which were cultured in 100 mL shaking flask. The harvested wet bacteria pellets were resuspended with 3 mL TE buffer (pH7.5). Then, TrxR1 were purified by ADP Sepharose affinity chromatography (see Fig. 4. for Coomassie-stained SDS-PAGE analyses). a Except that “-IPTG” means no IPTG, all the others were added with IPTG at a final concentration of 0.5 mmol/L.b The protein concentrations were evaluated using the Bradford method with BSA as protein reference. c Rat TrxR1 concentrations were measured at 463nm via FAD concentration determination (table not show). TrxR1 activities were measured by using the DTNB reduction assay. Each group of data was measured three times and the average value was shown in the table. | ||||||
表选项
这些结果也表明,大肠杆菌tRNASec骨架上的G18和G19基因在进化上是保守的,它们可能是维持大肠杆菌三级结构的关键核苷酸位点,我们通过突变tRNA骨架上的其他位点来寻找高活性的硒蛋白或许可以实现。
2.4 基于多点突变tRNASec的TrxR1制备与纯化 将a26、b7、b1接种到1 L锥形瓶中诱导表达TrxR1,依次经过2', 5' ADP-Sepharose亲和层析、超滤浓缩和凝胶过滤进行纯化,最后利用DTNB反应检测TrxR1纯酶的比活力。凝胶过滤后的TrxR1样品进行考马斯亮蓝染色分析,在55 kDa处可见明显目的条带。
纯化结果如表 5所示,经过凝胶过滤的TrxR1总活力下降较少,比活力上升。其中,a26的接近野生型tRNASec共表达的TrxR1比活力的一半,为8.6 U/mg;b7为6.21 U/mg;b1比活力最低,与之前结果较为一致,说明深孔板筛选鉴定的方案是可行的,即:首先构建特异位点突变体,利用24深孔板表达硒蛋白TrxR1,通过DTNB分析初步筛出酶活相对较高的tRNASec突变,最后利用亲和层析、凝胶过滤纯化对应的TrxR1,计算蛋白产量和比活力,对初始预判进行反馈修订。
表 5. TrxR1蛋白产量与比活力总结 Table 5. The yields and specific activities for TrxR1
| Mutation sitesa | ADP Sepharoseb | Sephacryl S-300c | ||||||
| mg | U/mgd | U | mg | U/mgd | U | |||
| W.T. | — | 14.99 | 6.30 | 94.41 | 4.63 | 19.10 | 88.51 | |
| a26 | U20A; A21G | 8.18 | 4.21 | 34.43 | 1.85 | 8.60 | 15.90 | |
| b7 | G64U; U65C | 17.04 | 1.16 | 19.73 | 2.88 | 6.21 | 17.88 | |
| b1 | U63C; G64A | 23.17 | 0.41 | 9.55 | 2.2 | 1.01 | 2.21 | |
| All TrxR1 proteins produced with tRNASec variants listed above were expressed in 1 L shaking flask and subsequently purified by affinity chromatography and gel filtration accordingly. a “—” means that there is no mutation in tRNA, and the others are the sites of the mutation in E. coli tRNASec.b TrxR1 were purified by 2, 5 ADP-Sepharose only. c TrxR1 were purified by 2, 5 ADP-Sepharose and then concentrated and finally purified by gel filtration. d Specific activity were determined in the standard DTNB reduction assay. | ||||||||
表选项
3 讨论 在原核生物系统中,tRNA依次与SerRS、SelA和SelB相互作用,获得UGA精准引导,从而使Sec掺入多肽链中,最终生成硒蛋白。当然,少数情况下UAG密码子也可以编码Sec产生硒蛋白[26]。但是,终止密码子UGA或UAG都能与释放因子结合,翻译过程终止,从而形成删减化硒蛋白[26-27]。提高掺硒效率、规避翻译终止是本研究主要目的。
在本研究中,大肠杆菌tRNASec的G18和G19位点的所有突变体生产酶活力远低于野生型的,说明G18和G19位点可能是E. coli tRNASec骨架上的关键核苷酸位点。2013年,Yuzuru Itoh等认为Thermoanaerobacter tengcongensis tRNASec三级结构由tRNA D臂和T臂维持,包括G18:U55 bp,Y16:Y59 (Y是C或U),U20:G19:C56和C15:G20a: G48等碱基[15]。通过BLAST比对原核tRNASec序列,也可以看出tRNASec D臂上的G18和G19位点序列保守。由此推断,E. coli tRNASec骨架上的G18、G19位点可能是维持tRNA三级结构的关键核苷酸位点,改变这两个位点,不利于tRNA与SelA或SelB结合,这或许为我们寻找掺硒效率高的硒蛋白提供了方向。这也说明了E. coli tRNASec具有序列可变性,一些关键核苷酸位点的改变,可能使得tRNASec减少和SelA、SelB相互作用的活性,从而严重影响TrxR1发挥功能,下一步通过进行蛋白质与核酸之间的相互作用来验证。与此结果类似,Tetsu M. Ishii发现tRNASec的D臂的A14:U21和C15:G20能够维持tRNA结构稳定性,然而并不在tRNA功能中起特异性作用[28]。我们发现改变tRNASec可以影响掺硒效率,进而发现G18和G19可能是大肠杆菌tRNASec的关键核苷酸位点,这为解决硒蛋白掺硒效率低、产量低提供了新思路。在后期研究中,我们先筛选出高酶活的突变体,然后对TrxR1功能进行性质表征。
定点突变是基于PCR技术向质粒目的DNA引入突变,根据突变位点的数量,可以分为单点突变和多点突变,一般用于有效提升DNA所表达的目的蛋白的性质表征[29]。然而,目前人们并未构建有完整的E. coli tRNASec与SelA、E. coli tRNASec与SelB晶体结构,这将不利于研究E. coli tRNASec对TrxR1掺硒效率的影响。在本研究中,我们以大鼠TrxR1硒蛋白为模型,通过对大肠杆菌tRNASec中一些可能与SelA/SelB结合的位点进行定点突变,寻找大肠杆菌tRNASec的关键核苷酸位点。我们发现改变大肠杆菌tRNASec的序列可以改变TrxR1硒蛋白的酶活。这与Christian Baron等[16]的早期结果类似,他们将E. coli tRNASec D臂上的G8、U9和U14位点进行碱基替换,FDHH、FDHN重组硒蛋白的比活力有所下降。这提示我们,通过改造tRNA寻找高品质硒蛋白的方法是可行的。同理,我们可以通过突变SECIS元件,寻找目标硒蛋白,目前在本实验室正在进行中。
在筛选方法上,我们先利用24深孔板进行粗筛,然后对酶活相对较高的TrxR1进行纯化。从后续的实验可以看出,在24深孔板或100 mL锥形瓶中共表达pSUABC'突变体所表达的TrxR1酶活变化趋势一致,这有效地减少了筛选的工作量。在之后的实验中,我们将进一步扩大突变范围,有望获得高品质硒蛋白。
在本研究中,我们根据RMA26和RMB7突变体的序列信息及测序结果,利用Secmarker (http://secmarker.crg.cat)在线预测tRNASec的二级结构[25],之后再利用RNAComposer (http://rnacomposer.ibch.poznan.pl/)在线预测RMA26和RMB7的空间结构,结果如图 6所示。RMA26和RMB7分别在骨架上突变了U20/A21和G64/U65,从图 6可以看出,新产生的tRNASec一级结构不同,导致它们的二级结构改变,在tRNA三级结构上造成差异,可能影响到与SelA和SelB的识别与结合,这对高效掺硒产生了影响。在后期研究中,我们将致力于筛选出高酶活的tRNASec突变体,精细研究tRNA新序列结构与掺硒元件的互作对TrxR1掺硒效率的改善。
 |
| 图 6 本研究中部分tRNASec突变体的二级结构和三级结构预测 Figure 6 Predicted secondary and tertiary structures of tRNASec mutants in this study. (A) Predicted secondary structure of RMA26 (U20A & A21G). (B) Predicted secondary structure of RMB7 (G64U & U65C). (C) Predicted structure of RMA26 (U20A & A21G). (D) Predicted structure of RMB7 (G64U & U65C). |
| 图选项 |
基于硒蛋白在大肠杆菌中的合成机制,我们在本研究中聚焦tRNASec进行位点突变和结构改造。大肠杆菌tRNASec的关键核苷酸位点参与调控Sec掺入及硒蛋白合成,这将揭示tRNASec与SelA/SelB之间的相互识别和调控机制。同时,我们为工程化改造tRNA合成全长硒蛋白提供了新方法。必须指出的是,我们需要扩大定点突变的范围,以便于寻找到适宜的新型tRNASec高活性的硒蛋白。本研究对优化大肠杆菌硒蛋白合成系统以获得高品质硒蛋白将提供有益参考。
References
| [1] | Avery JC, Hoffmann PR. Selenium, selenoproteins, and immunity. Nutrients, 2018, 10(9): 1203. DOI:10.3390/nu10091203 |
| [2] | Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. Selenium and selenocysteine:roles in cancer, health, and development. Trends in Biochemical Sciences, 2014, 39(3): 112-120. |
| [3] | Mousa R, Dardashti RN, Metanis N. Selenium and selenocysteine in protein chemistry. Angewandte Chemie International Edition in English, 2017, 56(50): 15818-15827. DOI:10.1002/anie.201706876 |
| [4] | Gao YH, Zhang JL, Huang XD, Zhang GX. Glutathione peroxidase 1, selenoprotein K, and selenoprotein H may play important roles in chicken testes in response to selenium deficiency. Biological Trace Element Research, 2017, 179(2): 271-276. DOI:10.1007/s12011-017-0953-y |
| [5] | Hoffman KS, Crnkovi? A, S?ll D. Versatility of synthetic tRNAs in genetic code expansion. Genes, 2018, 9(11): 537. DOI:10.3390/genes9110537 |
| [6] | Xu JQ, Croitoru V, Rutishauser D, Cheng Q, Arnér ESJ. Wobble decoding by the Escherichia coli selenocysteine insertion machinery. Nucleic Acids Research, 2013, 41(21): 9800-9811. DOI:10.1093/nar/gkt764 |
| [7] | Wang CY, Guo Y, Tian QN, Jia Q, Gao YZ, Zhang QF, Zhou C, Xie W. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Research, 2015, 43(21): 10534-10545. |
| [8] | Fu X, S?ll D, Sevostyanova A. Challenges of site-specific selenocysteine incorporation into proteins by Escherichia coli. RNA Biology, 2018, 15(4/5): 461-470. |
| [9] | Aldag C, Br?cker MJ, Hohn MJ, Prat L, Hammond G, Plummer A, S?ll D. Rewiring translation for elongation factor Tu-dependent selenocysteine incorporation. Angewandte Chemie International Edition in English, 2013, 52(5): 1441-1445. DOI:10.1002/anie.201207567 |
| [10] | Fan ZL, Song J, Guan TC, Lv XX, Wei JY. Efficient expression of glutathione peroxidase with chimeric tRNA in amber-less Escherichia coli. ACS Synthetic Biology, 2018, 7(1): 249-257. DOI:10.1021/acssynbio.7b00290 |
| [11] | Miller C, Br?cker MJ, Prat L, Ip K, Chirathivat N, Feiock A, Veszprémi M, S?ll D. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Letters, 2015, 589(17): 2194-2199. DOI:10.1016/j.febslet.2015.06.039 |
| [12] | Mukai T, Vargas-Rodriguez O, Englert M, Tripp HJ, Ivanova NN, Rubin EM, Kyrpides NC, S?ll D. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Research, 2017, 45(5): 2776-2785. |
| [13] | Sturchler C, Westhof E, Carbon P, Krol A. Unique secondary and tertiary structural features of the eucaryotic selenocysteine tRNASec. Nucleic Acids Research, 1993, 21(5): 1073-1079. DOI:10.1093/nar/21.5.1073 |
| [14] | Baron C, Westhof E, B?ck A, Giegé R. Solution structure of selenocysteine-inserting tRNASec from Escherichia coli:Comparison with canonical tRNASer. Journal of Molecular Biology, 1993, 231(2): 274-292. |
| [15] | Itoh Y, Br?cker MJ, Sekine S, Hammond G, Suetsugu S, S?ll D, Yokoyama S. Decameric SelA·tRNASec ring structure reveals mechanism of bacterial selenocysteine formation. Science, 2013, 340(6128): 75-78. |
| [16] | Itoh Y, Sekine SI, Yokoyama S. Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB. Nucleic Acids Research, 2015, 43(18): 9028-9038. DOI:10.1093/nar/gkv833 |
| [17] | Tormay P, Sawers A, B?ck A. Role of stoichiometry between mRNA, translation factor SelB and selenocysteyl-tRNA in selenoprotein synthesis. Molecular Microbiology, 1996, 21(6): 1253-1259. DOI:10.1046/j.1365-2958.1996.881450.x |
| [18] | Arnér ESJ. Recombinant expression of mammalian selenocysteine-containing thioredoxin reductase and other selenoproteins in Escherichia coli. Methods in Enzymology, 2002, 347: 226-235. DOI:10.1016/S0076-6879(02)47022-X |
| [19] | Prast-Nielsen S, Dexheimer TS, Schultz L, Stafford WC, Cheng Q, Xu JQ, Jadhav A, Arnér ESJ, Simeonov A. Inhibition of thioredoxin reductase 1 by porphyrins and other small molecules identified by a high-throughput screening assay. Free Radical Biology and Medicine, 2011, 50(9): 1114-1123. DOI:10.1016/j.freeradbiomed.2011.01.020 |
| [20] | Peng XX, Xu JQ, Arnér ESJ. Thiophosphate and selenite conversely modulate cell death induced by glutathione depletion or cisplatin:effects related to activity and Sec contents of thioredoxin reductase. Biochemical Journal, 2012, 447(1): 167-174. |
| [21] | Dominy CN, Andrews DW. Site-directed mutagenesis by inverse PCR//Casali N, Preston A. E. coli Plasmid Vectors. Totowa, NJ: Humana Press, 2003: 209-223. |
| [22] | Hsieh PC, Vaisvila R. Protein engineering: single or multiple site-directed mutagenesis//Samuelson JC. Enzyme Engineering: Methods and Protocols. Totowa, NJ: Humana Press, 2013: 173-186. |
| [23] | Xu JQ, Arnér ESJ. Pyrroloquinoline quinone modulates the kinetic parameters of the mammalian selenoprotein thioredoxin reductase 1 and is an inhibitor of glutathione reductase. Biochemical Pharmacology, 2012, 83(6): 815-820. DOI:10.1016/j.bcp.2011.12.028 |
| [24] | Xu JQ, Cheng Q, Arnér ESJ. Details in the catalytic mechanism of mammalian thioredoxin reductase 1 revealed using point mutations and juglone-coupled enzyme activities. Free Radical Biology and Medicine, 2016, 94: 110-120. DOI:10.1016/j.freeradbiomed.2016.02.013 |
| [25] | Santesmasses D, Mariotti M, Guigó R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Computational Biology, 2017, 13(2): e1005383. |
| [26] | Turanov AA, Xu XM, Carlson BA, Yoo MH, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Advances in Nutrition, 2011, 2(2): 122-128. DOI:10.3945/an.110.000265 |
| [27] | Carlson BA, Lee BJ, Tsuji PA, Tobe R, Park JM, Schweizer U, Gladyshev VN, Hatfield DL. Selenocysteine tRNA[Ser]Sec: From nonsense suppressor tRNA to the quintessential constituent in selenoprotein biosynthesis//Hatfield DL, Schweizer U, Tsuji PA, Gladyshev VN. Selenium. Cham: Springer, 2016: 3-12. |
| [28] | Ishii TM, Kotlova N, Tapsoba F, Steinberg SV. The long D-stem of the selenocysteine tRNA provides resilience at the expense of maximal function. The Journal of Biological Chemistry, 2013, 288(19): 13337-13344. DOI:10.1074/jbc.M112.434704 |
| [29] | Ling MM, Robinson BH. Approaches to DNA mutagenesis:an overview. Analytical Biochemistry, 1997, 254(2): 157-178. DOI:10.1006/abio.1997.2428 |
