芦晨阳1,2, 吴杭3, 苏秀榕1

 , 白林泉2
, 白林泉2 1.宁波大学海洋学院, 浙江 宁波 315211;
2.上海交通大学微生物代谢国家重点实验室, 上海 200240;
3.安徽大学生命科学学院, 安徽 合肥 230029
收稿日期:2016-10-04;修回日期:2016-12-02;网络出版日期:2016-12-30
基金项目:国家自然科学基金(31070070)
*通信作者:苏秀榕, Tel:+86-574-87608368;E-mail:suxiurong@nbu.edu.cn
摘要: [目的]测试大片段删减低转录区域对菌体生长和井冈霉素产量的影响。[方法]通过转录组分析,选择染色体上连续的基因低转录区域进行大片段缺失,通过Cre-loxP位点特异性重组得到1.2 Mb片段缺失突变株LCY-4。HPLC检测缺失株井冈霉素产量的变化,并测定干重绘制生长曲线。[结果]通过转录组分析,我们在井冈霉素高产菌株TL01染色体左侧末端发现了1.9 Mb的连续基因低转录区,使用Cre-loxP系统对其中的1.2 Mb区域进行大片段缺失,成功得到了1.2 Mb缺失突变株LCY-4。和出发菌株TL01相比,缺失突变株LCY-4中井冈霉素发酵产量基本保持不变,生物量有显著提高,最高增幅达到44%。[结论]1.2 Mb区域的成功缺失,意味着基于转录组分析寻找连续的基因低转录区域并加以缺失的策略的可行性。1.2 Mb片段缺失对菌体生物量积累具有明显促进作用,为后续将其开发成氨基环醇类药物异源表达的通用高产宿主奠定了基础。
关键词: 井冈霉素 转录组 Cre-loxP系统 大片段缺失
Effect of transcriptome-based large region deletion on validamycin A overproduction by Streptomyces hygroscopicus var. jinggangensis TL01
Chenyang Lu1,2, Hang Wu3, Xiurong Su1

 , Linquan Bai2
, Linquan Bai2 1.School of Marine Sciences, Ningbo University, Ningbo 315211, Zhejiang Province, China;
2.State Key Laboratory of Microbial Metabolism, Shanghai Jiao Tong University, Shanghai 200240, China;
3.School of Life Sciences, Anhui University, Hefei 230029, Anhui Province, China
Received 4 October 2016; Revised 2 December 2016; Published online 30 December 2016
*Corresponding author: Xiurong Su, Tel:+86-574-87608368;E-mail:suxiurong@nbu.edu.cn
Supported by the National Natural Science Foundation of China (31070070)
Abstract: [Objective]The effect of continuous low-expression region on validamycin A titer and biomass was evaluated through gene deletion experiments.[Methods]The deletion boundaries of large chromosome region were identified via transcriptome analysis. Meanwhile, deletion mutant was obtained via Cre-loxP system, titer of validamycin A was detected by HPLC and, biomass was represented by dry cell weight.[Results]The 1.2 Mb large region identified via transcriptome analysis was successfully deleted by Cre-loxP system. In comparison with parent strain TL01, deletion of 1.2 Mb region increased the biomass by 44% and showed no effect on validamycin A titer.[Conclusion]The strategy developed here can be applied to identify deletion boundaries of large chromosome region, and mutant of 1.2 Mb region deletion showed potential as host for heterologous expression of aminocyclitol biosynthetic gene clusters.
Key words: validamycin A transcriptome Cre-loxP system large region deletion
农用抗生素井冈霉素属于C7N氨基环醇类化合物,可高效防治水稻纹枯病,广泛应用于亚洲的水稻产区[1]。同时,井冈霉素生物合成中间产物可用于生产治疗Ⅱ型糖尿病的药物阿卡波糖和伏格列波糖,具有极高的经济价值[2]。2005年,井冈霉素生物合成基因簇被克隆,随后通过同位素标记、基因缺失和体外酶促等实验,证明其合成前体是来源于磷酸戊糖途径的7-磷酸景天庚酮糖、UDP-葡萄糖和谷氨酸[3-4]。2012年,井冈霉素野生型菌株吸水链霉菌井冈变种5008 (Streptomyces hygroscopicus var. jinggangensis 5008) 及其衍生的高产菌株TL01 (S. hygroscopicus var. jinggangensis TL01) 全基因组被测定,通过多组学手段对井冈霉素高产机理进行了详尽的阐述[4]。
5008及其衍生的高产菌株TL01基因组大小分别为10.1 Mb和9.84 Mb。与天蓝色链霉菌[5](S. coelicolor 8.67 Mb)、阿维链霉菌[6](S. avermitilis 9.03 Mb)、灰色链霉菌[7](S. griseus 8.55 Mb)、棒状链霉菌[8](S. clavuligerus 6.76 Mb)、白色链霉菌[9](S. albus 8.38 Mb)相比,井冈霉素产生菌具有较大的基因组规模,这可能导致了一些能量、前体和还原力的浪费,阻碍了抗生素产量的进一步提高。同时,前期的实验证明,大片段删减有利于减少前体、能量和还原力在复制、转录和翻译过程中的消耗,同时可以降低基因组的复杂性,提高菌株的发酵性能[10-12]。但是,针对全新的菌株,目前尚缺乏有效的策略来精确确定可删减的大片段区域,这限制了大片段删减技术的推广应用。
同源重组双交换技术是在链霉菌中使用最广泛的基因删减技术[13-14],但是在对大片段区域进行删减的过程中,研究者发现其往往会导致不规则敲除现象的发生[10]。同源臂长度的增加对降低不规则敲除现象出现的频率没有明显帮助。以Cre-loxP为代表的位点特异性重组系统可以很好地解决这一问题,在酵母、植物、果蝇和哺乳动物遗传操作方面具有广泛的使用[15-17]。Fedoryshyn和Lopatniuk等研究者利用Cre-loxP系统在阿维链霉菌和加纳链霉菌中成功实现了大范围的基因组删减[18-19]。
本研究中,通过对井冈霉素野生型菌株5008及其衍生的高产菌株TL01的全局性转录谱分析,我们发现在TL01染色体左侧末端存在1.9 Mb的连续基因低转录区域,判断其可能是生长非必需区域。随后,使用Cre-loxP特异性重组系统对该区域中1.2 Mb片段进行缺失,考察了1.2 Mb片段缺失对井冈霉素产量和菌体生物量的影响,并对突变株相关表型产生的机理进行了初步分析。
1 材料和方法 1.1 材料
1.1.1 菌种、质粒及引物: 本研究所用的菌种、质粒及引物见表 1。 表 1. 本研究所用的菌株、质粒及引物 Table 1. Strains, plasmids and primers used in this study
| Strains, plasmids and primers | Related characters and sequences | Sources |
| S. hygroscopicus var. jinggangensis | ||
| 5008 | Wild-type producer of validamycin A | Shanghai Jiao Tong University |
| TL01 | High-yielding producer of validamycin A | Zhejiang Tonglu Huifeng Biosciences Co., Ltd. |
| LCY-1 | Replacement of 452, 301-453, 069-bp region with loxP+aac3(Ⅳ) cassette in TL01 (AprR) | This study |
| LCY-2 | Replacement of 1, 610, 359-1, 610, 972-bp region with neo +loxP cassette in LCY-1 (AprRKanR) | This study |
| LCY-3 | Over-expression of the plasmid pLQ102 carrying gene cre in LCY-2 (AprRKanRThioR) | This study |
| LCY-4 | Deletion of 1.2 Mb region in TL01 (AprSKanSThioS) | This study |
| E. coli | ||
| DH10B | F′(traΔ36 lacIq proAB lacZΔM15) rpsL (strR)thr leu endA thi-1 lacY galK galT ara tonA tsx dcm dam supE44 Δ(lac-proAB)Δ(mcrC-mrr)102::Tn10(tetR) | GIBCO BRL |
| ET12567(pUZ8002) | recE dcm-dam-hsdS Cmr Tetr Strr Kmr | [20] |
| Plasmids | ||
| pBluescript Ⅱ SK(+) | Bla lacZ orif1 | Stratagene |
| pJTU1278 | Bla tsr lacZ oriT oripIJ101 oriColE1 | [21] |
| pUWLCRE | Plasmid with the gene cre | [18] |
| pLQ100 | Construct with region (452, 301-453, 069 bp) replaced by loxP+aac3(Ⅳ) cassette | This study |
| pLQ101 | Construct with region (1, 610, 359-1, 610, 972 bp) replaced by neo+loxP cassette | This study |
| pLQ102 | Plasmid pJTU1278 carrying gene cre under the control of PermE* promoter | This study |
| Primers | 5′→3′ | |
| loxP+aac3(Ⅳ)-F-Bgl Ⅱ | ATATAAGATCTATAACTTCGTATAGCATACATTATACGAAGTTATGATATCGAATTCCCCAAT GT (loxP site in bold) | |
| loxP+aac3(Ⅳ)-R-Hind Ⅲ | ATATAAAGCTTGCATGCCGGTCGACTCTAGA | |
| Arm1-F-Spe Ⅰ | ATATAACTAGTGGCGATGATGATGCTCTC3 | |
| Arm1-R-Bgl Ⅱ | ATATAAGATCTCAGCGACCAGATGAAGAC | |
| Arm2-F-Hind Ⅲ | ATATAAAGCTTGCAGGTACTCGCTCTTGT | |
| Arm2-R-KpnⅠ | TCCTACGTCACCGTGTTG | |
| loxP-A-F | CTTGACAGCGTTGTCGAG | |
| loxP-A-R | CGAAGTACAAGAGCGAGTA | |
| neo+loxP-F-BamH Ⅰ | ATATAGGATCCCACGCTGCCGCAAGCACTCA | |
| neo+loxP-R-Hind Ⅲ | TATAAAGCTTATAACTTCGTATAATGTATGCTATACGAAGTTATGTCCCGCTCAGAAGAACT CG (loxP site in bold) | |
| Arm3-F-Spe Ⅰ | ATATAACTAGTTGGAAGGTGATGTCGATGAG | |
| Arm3-R-BamH Ⅰ | ATATAGGATCCGCTCGGTCAAGACCAATCTC | |
| Arm4-F-Hind Ⅲ | ATATAAATCTTTGGTCCATGTAGTCGTTGTA | |
| Arm4-R-Kpn Ⅰ | ATATAGGTACCGAGTTCTTCTCACGCCACTA | |
| loxP-B-F | CACAGGACGGTCTTGATC | |
| loxP-B-R | CTTCACAGGACTGAGTTACA | |
| Cre-F-Nde Ⅰ | ATATATCATATGTCCAACCTGCTGACCGT | |
| Cre-R-EcoR Ⅰ | ATATATGAATTCTCAGTCGCCGTCTTCCAGCA | |
| Cre-RT-F | GGAAGAACCTGATGGACAT | |
| Cre-RT-R | GGAGGTACAGGAGGTAGTC | |
| loxP-C-F | TCTCCAGCAGCGAGTTGA | |
| loxP-C-R | CGATTCTCCAGCATCCTTATG | |
| SHJG0599-F | CCTACGACGGGTTGTGGGAACG | |
| SHJG0599-R | GCAGCAGCATGTCGCGGAAGA | |
| SHJG1058-F | GGTGGTCATCCCGTCCCTGTC | |
| SHJG1058-R | CGAGGGTGCCGTGGAAGAAG | |
| SHJG1338-F | ACCGGCTCGCCCTGTTGATG | |
| SHJG1338-R | TGACGCTGATGCCGTACTTGACG | |
表选项
1.1.2 培养基: 大肠杆菌培养采用LB (Luria-Bertani)和LA培养基;链霉菌培养采用TSBY液体培养基:胰胨豆汤粉30 g,蔗糖103 g,酵母提取物10 g,蒸馏水1000 mL;黄豆饼粉浸汁琼脂(SFM):黄豆饼粉20 g,甘露醇20 g,琼脂20 g,蒸馏水1000 mL;YMG培养基:酵母提取物4 g,麦芽提取物10 g,葡萄糖4 g,蒸馏水1000 mL;井冈霉素发酵培养基:大米粉95 g,花生粉18 g,K2HPO4 0.7 g,NaCl 1.4 g,CaCO3 0.6 g,蒸馏水1000 mL[22]。
1.1.3 主要试剂: 实验中所用抗生素购自Sigma公司;连接酶和KOD高保真DNA聚合酶购自TOYOBO公司;PCR产物回收试剂盒购自Omega公司;限制性内切酶购自Fermentas公司;RNA快速提取试剂盒购自赛百盛公司。1.2 loxP位点插入质粒构建 以TL01基因组DNA为模板,用同源臂扩增引物扩增得到预测大小的同源臂片段。以pJTU472或者SuperCos1为模板,用loxP片段引物扩增得到预测大小的loxP+aac3(Ⅳ)或neo+loxP片段。三片段连接载体pJTU1278后得到的质粒验证正确即为loxP位点插入质粒。
1.3 Cre蛋白表达质粒构建 以质粒pUWLCRE为模板,使用cre基因扩增引物得到基因片段。此片段经Nde Ⅰ/EcoR Ⅰ酶切之后连接到载体pJTU968,随后将验证正确的质粒用Mun Ⅰ/EcoR Ⅰ双酶切处理,将其中1.2 kb的片段连接到用EcoR Ⅰ单酶切处理后的载体pJTU1278,得到目的质粒pLQ102。
1.4 接合转移及突变株筛选 将pLQ100转入大肠杆菌ET12567 (pUZ8002) 中,通过大肠杆菌-链霉菌间的接合转移导入井冈霉素高产菌株TL01中。16 h后用含安普霉素和甲氧苄啶两种抗生素的无菌水覆盖,待SFM平板吹干后,倒置培养3-4 d可以观察到接合子。将接合转移子划线接种至含上述两种抗生素的SFM平板上30 ℃培养。用验证引物验证接合转移子是单交换菌株后,将正确的单交换菌株进行松弛培养,收集孢子进行梯度稀释,得到单菌落。随后用PCR扩增方法筛选得到双交换突变株。
1.5 cre基因转录水平检测 在添加安普霉素、卡那霉素和硫链丝菌素的YMG培养基中培养LCY-3 48 h之后,收集菌丝体抽提RNA。将抽提的RNA用DNase-I消化4 h之后作为模板,使用引物cre-RT-F/cre-RT-R进行PCR验证,以确定基因组DNA消化干净。随后,对RNA进行逆转录,以得到的cDNA作为模板、cre-RT-F/cre-RT-R为引物进行PCR验证,检测是否有cre基因转录产生的RNA。
1.6 1.2 Mb片段缺失突变株的筛选和验证 将同时携带2个loxP位点和cre基因的突变株在无抗TSBY培养基中转接2轮,随后涂布在无抗的SFM培养基中进行松弛培养,收集孢子进行梯度稀释,得到单菌落。筛选对安普霉素和卡那霉素均敏感的单克隆,使用验证引物进行验证,正确即为1.2 Mb片段缺失突变株,命名为LCY-4。
1.7 井冈霉素发酵及含量检测 将得到的突变株LCY-4与出发菌株TL01在种子培养基TSBY中30 ℃、220 r/min振荡培养36-48 h后,按体积比1:10接入发酵培养基,37 ℃、220 r/min振荡发酵5 d。每隔24 h准确量取1 mL的发酵液,采用高效液相色谱(HPLC)法测定,操作如下:将发酵液12000 r/min离心15 min取上清,用0.2 μm一次性水相过滤器过滤,过滤后的样品稀释10倍,用HPLC检测井冈霉素产量。所用仪器为安捷伦1200系列高效液相色谱仪,色谱柱为Agilent公司的ZORBAX SB-C18柱(3.5 μm,2.1×150 mm),流动相为98%的0.5 mmol/L磷酸盐缓冲液和2%甲醇,流速为0.1 mL/min,柱温为室温,检测波长为210 nm。每针进样量为5 μL,检测时间为15 min。根据标准曲线,计算出井冈霉素产量。
1.8 菌体干重测定 菌体干重测定采用YMG培养基,37 ℃、220 r/min振荡发酵4 d,每隔24 h准确量取1 mL的菌液放于已称重的离心管中,8000 r/min离心弃上清,用无菌水清洗1-2遍,以除去残留的培养基。将离心管放在70 ℃烘箱中,烘至恒重称量,计算菌体干重。
2 结果和分析 2.1 基于转录谱分析,确定井冈霉素产生菌中“非必需区域” 通过转录组芯片分析(Microarray analysis),我们比较了井冈霉素产生菌野生型菌株5008及其衍生的高产菌株TL01在高产培养基条件下全局性的基因表达谱(P < 0.05) (图 1)。从染色体左侧末端开始接近1.9 Mb的区域内,只有井冈霉素合成基因簇和一个Ⅱ型PKS生物合成基因簇是转录上调,其余都是转录下调的(图 1)。这种染色体上区域性的转录谱变化,暗示着TL01上这段低转录区域可能是生长非必需的,可以进行缺失来减少基因组规模、降低基因组复杂度。为了保留井冈霉素生物合成基因簇,我们选择了图 1所示1.2 Mb的区域进行后续的缺失实验。
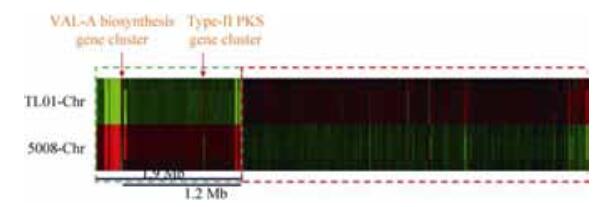 |
| 图 1 在高产培养基条件下5008与TL01全局性的基因表达谱(P < 0.05) Figure 1 Global gene expression profiling of 5008 and TL01 in the high-yielding medium. P < 0.05. |
| 图选项 |
2.2 利用Cre-loxP特异性重组完成1.2 Mb片段缺失 我们采用Cre-loxP特异性重组对大片段进行缺失(图 2)。具体流程如下:(1) 在1.2 Mb区域的左侧边界和右侧边界分别插入loxP+aac3(Ⅳ)和neo+loxP片段,得到突变株LCY-2;(2) 在Cre蛋白介导下,2个同向的loxP位点之间发生片段环出,实现片段缺失,突变株LCY-4中保留一个loxP位点。
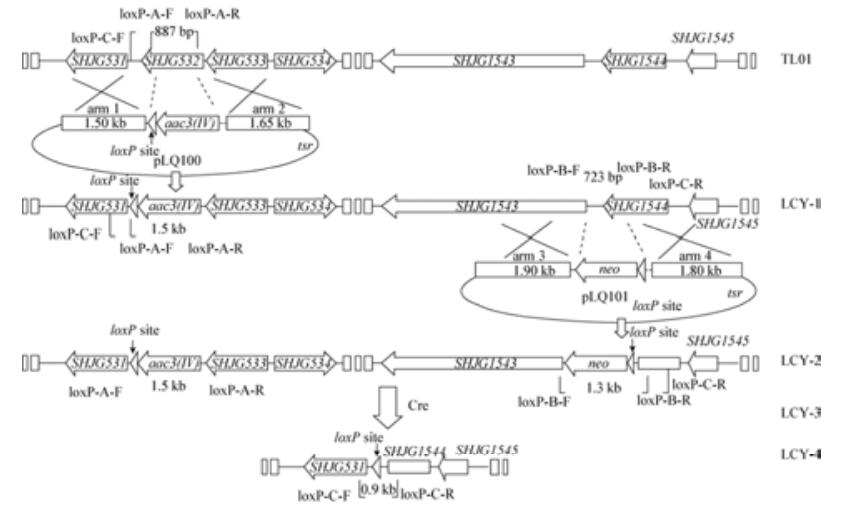 |
| 图 2 Cre-loxP特异性重组系统敲除1.2 Mb片段区域的示意图 Figure 2 Schematic representation of the deletion of 1.2 Mb region via Cre-loxP system. |
| 图选项 |
2.2.1 loxP+aac3(Ⅳ)片段定向插入1.2 Mb片段缺失边界左侧: 通过同源重组双交换的方式,将loxP+aac3(Ⅳ)片段定向插入染色体452 301-453 069 bp位置,得到的突变株命名为LCY-1 (图 2)。利用验证引物loxP-A-F/loxP-A-R对双交换菌株的基因型进行PCR扩增验证。以高产菌株TL01的总DNA为模板,扩增条带为0.90 kb;以缺失株LCY-1的总DNA为模板,扩增条带为1.60 kb,与预测一致(图 3-A)。
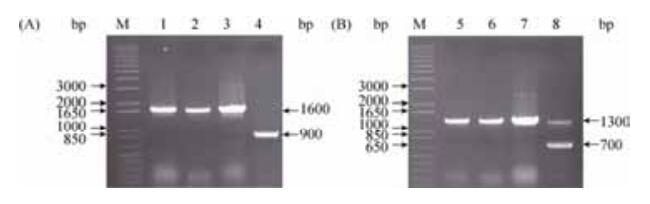 |
| 图 3 两个同向loxP位点通过同源重组插入1.2 Mb片段边界 Figure 3 Schematic representation of the insertion of two loxP sites in the boundaries of 1.2 Mb region. A: confirmation of LCY-1 by PCR amplification; B: confirmation of LCY-2 by PCR amplification. M: 1 kb ladder DNA marker. Lane 1-2: PCR product with the genomic DNA of mutant LCY-1 as template and loxP-A-F/loxP-A-R as primers. Lane 3: PCR product with pLQ100 as template and loxP-A-F/loxP-A-R as primers. Lane 4: PCR product with the genomic DNA of strain TL01 as template and loxP-A-F/loxP-A-R as primers. Lane 5-6: PCR product with the genomic DNA of mutant LCY-2 as template and loxP-B-F/loxP-B-R as primers. Lane 7: PCR product with pLQ101 as template and loxP-B-F/loxP-B-R as primers. Lane 8: PCR product with the genomic DNA of strain TL01 as template and loxP-B-F/loxP-B-R as primers. |
| 图选项 |
2.2.2 neo+loxP片段定向插入1.2 Mb片段缺失边界右侧: 通过同源重组双交换的方式,在突变株LCY-1的基础上,将neo+loxP片段定向插入染色体1610359-1610972 bp位置,得到的突变株命名为LCY-2 (图 2)。利用验证引物loxP-B-F/loxP-B-R对双交换菌株的基因型进行PCR扩增验证。以高产菌株TL01的总DNA为模板,扩增条带为0.70 kb;以缺失株LCY-2的总DNA为模板,扩增条带为1.30 kb,与预测一致(图 3-B)。
2.2.3 cre基因在LCY-3突变株中的表达: 为了检测突变株LCY-3中cre基因是否发生转录,我们将LCY-3中提取的RNA样品进行DNase I消化之后作为模板,使用引物cre-RT-F/cre-RT-R作为引物进行PCR扩增,由于DNA聚合酶不能使用RNA作为模板,因此无法得到150 bp的目标条带。之后以RNA逆转录得到的cDNA作为模板,使用cre-RT-F/cre-RT-R作为引物进行PCR扩增,得到150 bp的目标条带(图 4-A),说明在突变株LCY-3中cre基因已经转录。
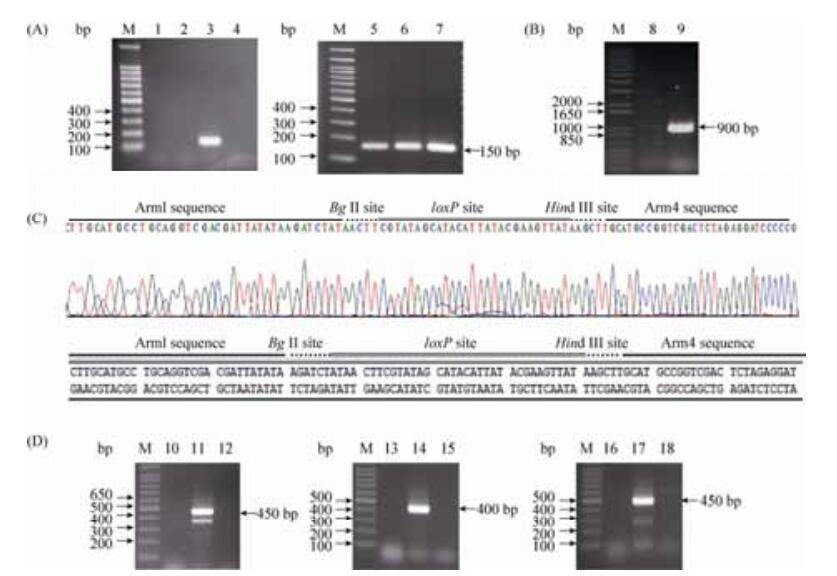 |
| 图 4 1.2 Mb片段缺失突变株筛选验证 Figure 4 Confirmation of mutant LCY-4 with 1.2 Mb region deletion. A: expression of gene cre via reverse-transcription PCR (RT-PCR); B: confirmation of LCY-4 by PCR amplification using primers loxP-C-F/loxP-C-R; C: sequencing and data analysis of PCR products; D: confirmation of LCY-4 by PCR amplification using primers designed in the 1.2 Mb large region. M: 1 kb ladder DNA marker. Lane 1-2: PCR product with the DNase I-digested RNA of mutant LCY-3 as template and cre-RT-F/cre-RT-R as primers. Lane 3 and 7: PCR product with pLQ102 as template and cre-RT-F/cre-RT-R as primers. Lane 4: PCR product with the genomic DNA of strain TL01 as template and cre-RT-F/cre-RT-R as primers. Lane 5-6: PCR product with the cDNA of mutant LCY-3 as template and cre-RT-F/cre-RT-R as primers. Lane 8: PCR product with the genomic DNA of strain TL01 as template and loxP-C-F/loxP-C-R as primers. Lane 9: PCR product with the genomic DNA of mutant LCY-4 as template and loxP-C-F/loxP-C-R as primers. Lane 10: PCR product with the genomic DNA of mutant LCY-4 as template and SHJG0599-C-F/SHJG0599-C-R as primers. Lane 11: PCR product with the genomic DNA of strain TL01 as template and SHJG0599-C-F/SHJG0599-C-R as primers. Lane 12: PCR product with the water as template and SHJG0599-C-F/SHJG0599-C-R as primers. Lane 13: PCR product with the genomic DNA of mutant LCY-4 as template and SHJG1058-C-F/SHJG1058-C-R as primers. Lane 14: PCR product with the genomic DNA of strain TL01 as template and SHJG1058-C-F/SHJG1058-C-R as primers. Lane 15: PCR product with the water as template and SHJG1058-C-F/SHJG1058-C-R as primers. Lane 16: PCR product with the genomic DNA of mutant LCY-4 as template and SHJG1338-C-F/SHJG1338-C-R as primers. Lane 17: PCR product with the genomic DNA of strain TL01 as template and SHJG1338-C-F/R as primers. Lane 18: PCR product with the water as template and SHJG1338-C-F/SHJG1338-C-R as primers. |
| 图选项 |
2.2.4 1.2 Mb缺失突变株LCY-4的筛选和验证: 首先,利用验证引物loxP-C-F/loxP-C-R对突变株进行PCR扩增验证(图 2)。以高产菌株TL01的总DNA为模板,由于目标条带太大(约1.16 Mb),无法得到目标条带;以突变株LCY-4的总DNA为模板,扩增条带0.90 kb (图 4-B)。对0.90 kb片段进行了测序和序列分析,发现这一段序列主要包含:同源臂1 (arm 1) 序列,Bgl Ⅱ酶切位点,loxP位点,EcoR Ⅴ酶切位点和同源臂4 (arm 4) 序列,测序结果和理论预测一致。同时,在缺失区域中随机选择3个基因(SHJG0599、SHJG1058和SHJG1338),设计内部引物进行PCR,以高产菌株TL01的总DNA为模板,扩增条带分别为0.45、0.40和0.45 kb;以缺失株LCY-4的总DNA为模板,由于此区域已经被缺失,均无法扩增到目标条带,与预测一致(图 4-D)。2.3 1.2 Mb片段缺失突变株LCY-4的发酵表型
2.3.1 1.2 Mb片段缺失不影响井冈霉素产量: 使用Cre-loxP特异性重组系统,顺利获得了1.2 Mb片段缺失的突变株LCY-4。将出发菌株TL01和LCY-4的3个平行株在相同条件下进行发酵,每组4个重复。和出发菌株相比,1.2 Mb缺失突变株LCY-4产素曲线没有明显变化,1.2 Mb片段缺失对井冈霉素产量提高没有促进作用(图 5-A)。
 |
| 图 5 1.2 Mb片段缺失对井冈霉素产量和菌体生物量的影响 Figure 5 Effects of 1.2 Mb region deletion on validamycin A production and cell growth. A: validamycin A production of TL01 and LCY-4; B: dry cell weight of TL01 and LCY-4. |
| 图选项 |
2.3.2 1.2 Mb片段缺失促进菌体生物量积累: 我们测定了出发菌株TL01和突变株LCY-4的生长曲线并进行比较,突变株LCY-4的生物量在发酵前24 h内有大幅度的提升,相比于出发菌株TL01上升了近44%,在48 h达到峰值,较出发菌株增加了26%。之后生物量在72 h趋近于出发菌株,但是随着发酵过程的进一步进行,在发酵96 h时突变株LCY-4的生物量重新回到高位,较出发菌株增加24% (图 5-B)。3 讨论 随着全基因组测序的发展,越来越多的链霉菌基因组信息被获取。由于基因组删减可以节约前体、能量和还原力,降低基因组的复杂度,因此在基因组改造中占有重要的位置[9, 23]。已报道的基因组删减策略主要集中在次级代谢产物生物合成基因簇敲除方面,通过对竞争基因簇的缺失来重新导向前体代谢流,促进目标产物的产量提高[9, 23-24]。但是对于基因组其他位置,如何有效界定非生长必需区域仍然是一个问题。在本文中,我们利用转录组的手段来寻找染色体上的连续基因低转录区域,考虑到连续的低转录区域(沉默区域)低效率参与转录、翻译及其他生化过程,但其在DNA复制过程中仍需要消耗大量的物质和能量,因此将其作为基因组删减的目标[25-26]。通过转录组芯片技术,我们发现在染色体左侧末端的1.9 Mb区域均为低转录区域,对其中井冈霉素生物合成基因簇右侧的1.2 Mb区域进行了缺失,成功得到了1.2 Mb缺失突变株LCY-4,证明了“基于转录组数据分析确定非生长必需区域”策略的可行性。
使用Cre-loxP系统可以非常高效精准地进行大片段缺失,但是缺失突变株中会残留一个loxP位点,对于后续采用该方法多个大片段的连续删减存在影响,可以采用Cre蛋白识别的突变loxP组合来克服该问题,重组完成之后残留的序列不被Cre蛋白识别[27]。
和出发菌株TL01相比,1.2 Mb片段缺失突变株在井冈霉素的产量上没有变化,但是生物量整体呈现增加的趋势。之所以井冈霉素产量没有增加,我们分析可能有以下几个原因:(1) 大片段删减后节约的能量、前体和还原力被用于生物量的积累,没有被井冈霉素生物合成途径有效利用;(2) 突变株LCY-4的发酵潜力在原高产培养基中无法体现,需要针对菌株特点重新设计优化培养基配方[28-29];(3) 高产菌TL01的井冈霉素产量已高达18-20 g/L,此时能量、前体、还原力的供应已不是其产量进一步提高的限制因子,尚存在其他限速因子[22]。
虽然1.2 Mb片段缺失突变株LCY-4没有促进井冈霉素产量的进一步增加,但其仍不失为一种优良的异源表达宿主,特别是大片段缺失之后菌株生物量明显增加,这符合现代发酵工艺对次级代谢产物生产菌株的要求[30-31]。此外,TL01和LCY-4中井冈霉素产量可以达到18-20 g/L,而游动放线菌SE50/110 (Actinoplanes sp. SE50/110) 中阿卡波糖产量为2-3 g/L,井冈霉素和阿卡波糖生物合成过程中共享部分的前体,因此后续可以考虑将此突变株开发成阿卡波糖等氨基环醇类抗生素的异源表达通用宿主。
References
| [1] | Iwasa T, Higashide E, Shibata M. Studies of validamycins, new antibiotics. 3. Bioassay methods for the determination of validamycin. The Journal of Antibiotics, 1971, 24(2): 114-118. DOI:10.7164/antibiotics.24.114 |
| [2] | Mahmud T, Lee S, Floss HG. The biosynthesis of acarbose and validamycin. The Chemical Record, 2001, 1(4): 300-310. DOI:10.1002/(ISSN)1528-0691 |
| [3] | Bai LQ, Li L, Xu H, Minagawa K, Yu Y, Zhang YR, Zhou XF, Floss HG, Mahmud T, Deng ZX. Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A. Chemistry & Biology, 2006, 13(4): 387-397. |
| [4] | Wu H, Qu S, Lu CY, Zheng HJ, Zhou XF, Bai LQ, Deng ZX. Genomic and transcriptomic insights into the thermo-regulated biosynthesis of validamycin in Streptomyces hygroscopicus 5008. BMC Genomics, 2012, 13(1): 337. DOI:10.1186/1471-2164-13-337 |
| [5] | Bentley SD, Chater KF, Cerde?o-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 2002, 417(6885): 141-147. DOI:10.1038/417141a |
| [6] | Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, ōmura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nature Biotechnology, 2003, 21(5): 526-531. DOI:10.1038/nbt820 |
| [7] | Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. Journal of Bacteriology, 2008, 190(11): 4050-4060. DOI:10.1128/JB.00204-08 |
| [8] | Song JY, Jeong H, Yu DS, Fischbach MA, Park HS, Kim JJ, Seo JS, Jensen SE, Oh TK, Lee KJ, Kim JF. Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. Journal of Bacteriology, 2010, 192(23): 6317-6318. DOI:10.1128/JB.00859-10 |
| [9] | Lu CY, Zhang XJ, Ming J, Bai LQ. Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metabolic Engineering, 2016, 35: 129-137. DOI:10.1016/j.ymben.2016.02.012 |
| [10] | Komatsu M, Uchiyama T, ōmura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(6): 2646-2651. DOI:10.1073/pnas.0914833107 |
| [11] | Liu ZQ, Xie YL, Zhang X, Hu XF, Li YS, Ding XZ, Xia LQ, Hu SB. Efficient construction of large genomic deletion in Agrobacterium tumefaciens by combination of Cre/loxP system and triple recombineering. Current Microbiology, 2016, 72(4): 465-472. DOI:10.1007/s00284-015-0977-5 |
| [12] | Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Müller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E. The sequence of a 1 1.8 Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biology and Evolution, 2010, 2: 212-224. DOI:10.1093/gbe/evq013 |
| [13] | Capecchi MR. Altering the genome by homologous recombination. Science, 1989, 244(4910): 1288-1292. DOI:10.1126/science.2660260 |
| [14] | Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromsome. Nature Biotechnology, 1997, 15(9): 859-865. DOI:10.1038/nbt0997-859 |
| [15] | Delneri D, Tomlin GC, Wixon JL, Hutter A, Sefton M, Louis EJ, Oliver SG. Exploring redundancy in the yeast genome:an improved strategy for use of the Cre-loxP system. Gene, 2000, 252(12): 127-135. |
| [16] | Li ZS, Xing AQ, Moon BP, Burgoyne SA, Guida AD, Liang HL, Lee C, Caster CS, Barton JE, Klein TM, Falco SC. A Cre/loxP-mediated self-activating gene excision system to produce marker gene free transgenic soybean plants. Plant Molecular Biology, 2007, 65(3): 329-341. DOI:10.1007/s11103-007-9223-2 |
| [17] | Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Research, 1995, 23(24): 5080-5081. DOI:10.1093/nar/23.24.5080 |
| [18] | Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A. Functional expression of the Cre recombinase in actinomycetes. Applied Microbiology and Biotechnology, 2008, 78(6): 1065-1070. DOI:10.1007/s00253-008-1382-9 |
| [19] | Lopatniuk M, Ostash B, Makitrynskyy R, Walker S, Luzhetskyy A, Fedorenko V. Testing the utility of site-specific recombinases for manipulations of genome of moenomycin producer Streptomyces ghanaensis ATCC14672. Journal of Applied Genetics, 2015, 56(4): 547-550. DOI:10.1007/s13353-015-0283-8 |
| [20] | Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor ?E is required for normal cell wall structure in Streptomyces coelicolor A3(2). Journal of Bacteriology, 1999, 181(1): 204-211. |
| [21] | He YL, Wang ZJ, Bai LQ, Liang JD, Zhou XF, Deng ZX. Two pHZ1358-derivative vectors for efficient gene knockout in Streptomyces. Journal of Microbiology and Biotechnology, 2010, 20(4): 678-682. DOI:10.4014/jmb |
| [22] | Zhou X, Wu H, Li Z, Zhou XF, Bai LQ, Deng ZX. Over-expression of UDP-glucose pyrophosphorylase increases validamycin A but decreases validoxylamine A production in Streptomyces hygroscopicus var. jinggangensis 5008. Metabolic Engineering, 2011, 13(6): 768-776. DOI:10.1016/j.ymben.2011.10.001 |
| [23] | Wei X, Liang YX, Zheng YH. Enhancement and selective production of oligomycin through inactivation of avermectin's starter unit in Streptomyces avermitilis. Biotechnology Letters, 2006, 28(12): 911-916. DOI:10.1007/s10529-006-9012-z |
| [24] | Schwientek P, Szczepanowski R, Rückert C, Kalinowski J, Klein A, Selber K, Wehmeier UF, Stoye J, Pühler A. The complete genome sequence of the acarbose producer Actinoplanes sp. SE50/110. BMC Genomics, 2012, 13: 112. DOI:10.1186/1471-2164-13-112 |
| [25] | Beacham IR. Silent genes in prokaryotes. FEMS Microbiology Letters, 1987, 46(4): 409-417. DOI:10.1111/fml.1987.46.issue-4 |
| [26] | Cornish-Bowden A, Cardenas ML. Silent genes given voice. Nature, 2001, 409(6820): 571. DOI:10.1038/35054646 |
| [27] | Arakawa H, Lodygin D, Buerstedde J M. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnology, 2001, 1: 7. DOI:10.1186/1472-6750-1-7 |
| [28] | Choi SY, Si JP, Kim WJ, Yang JE, Lee H, Shin J, Sang YL. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nature Biotechnology, 2016, 34(4): 435-440. DOI:10.1038/nbt.3485 |
| [29] | Kim SY, Lee J, Sang YL. Metabolic engineering of Corynebacterium glutamicum for the production of L-ornithine. Biotechnology and Bioengineering, 2015, 112(2): 416-421. DOI:10.1002/bit.v112.2 |
| [30] | Yuan YL, Guo XN, Zhang BR, Liu SG. Breeding of a high-biomass, iron-enriched yeast strain and its fermentation conditions. Industrial Microbiology, 2004, 34(4): 29-33. (in Chinese) 袁玉兰, 郭雪娜, 张博润, 刘世贵. 高生物量富铁酵母菌的选育及其发酵条件的研究. 工业微生物, 2004, 34(4): 29-33. |
| [31] | Zhao XL, Zhen YG, Wang LH, Yi XF. Aerobic fermentation culture medium of Saccharomyces cerevisiae. China Brewing, 2014, 33(7): 43-47. (in Chinese) 赵小丽, 甄玉国, 王兰惠, 易晓菲. 酿酒酵母有氧发酵培养基的研究. 中国酿造, 2014, 33(7): 43-47. DOI:10.11882/j.issn.0254-5071.2014.07.009 |
