姜南1,2, 刘卫1, 李岩1, 解志红1


1.中国科学院烟台海岸带研究所,山东 烟台 264003;
2.中国科学院大学,北京 100049
收稿日期:2015-10-27;修回日期:2015-12-02;网络出版日期:2015-12-21
基金项目:国家自然科学基金(31370108,31570063);中科院重点部署项目(KZZD-EW-14);山东省科技创新基金 (2014ZZCX07303);中科院****项目;烟台市科技发展计划(2013JH021)
*通信作者:Tel: +86-535-2109183; Fax: +86-535-2109000; E-mail: zhxie@yic.ac.cn
摘要: [目的]茎瘤固氮根瘤菌既可以与毛萼田菁共生固氮,又可以自生或作为内生菌在其他植物体内固氮。由于其具有3种生活状态及固氮能力,其感受外界信号的趋化系统应当更为复杂多样。目前对茎瘤固氮根瘤菌的趋化通路研究很少,因此我们将茎瘤固氮根瘤菌的趋化系统与其他已有研究的菌株相比较。[方法]基于NCBI蛋白数据库,利用BLAST程序对菌株ORS571及已公布全基因组序列的α变形菌门的其他菌种进行趋化基因簇的比较分析;基于Pfam蛋白数据库,利用HMMER3程序对甲基化受体蛋白序列进行比较分析。[结果]分析结果表明茎瘤固氮根瘤菌中有1条主要的趋化基因簇,其中所编码的甲基化酶CheR为非五肽依赖型;此外,该菌还具有43个甲基化受体蛋白基因,所编码的受体蛋白保守区段均由38个七肽单位组成。[结论]通过比较基因组学的分析可知茎瘤固氮根瘤菌与其他菌属相比趋化系统具有高度同源性,但同时存在自身的独特性,这一结论能够使我们更好的了解茎瘤固氮根瘤菌利用趋化系统适应环境的过程。
关键词: 趋化性 甲基化受体蛋白 茎瘤固氮根瘤菌
Comparative genomic and protein sequence analyses of the chemotaxis system of Azorhizobium caulinodans
Nan Jiang1,2, Wei Liu1, Yan Li1, Zhihong Xie1


1.Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, Shandong Province, China;
2.University of Chinese Academy of Sciences, Beijing 100049, China
Received 27 October 2015; Revised 02 December 2015; Published online 21 December 2015
*Corresponding author: Tel: +86-535-2109183; Fax: +86-535-2109000; E-mail: zhxie@yic.ac.cn
Supported by the National Natural Science Foundation of China (31370108, 31570063), by the Key Research Program of the Chinese Academy of Sciences (KZZD-EW-14), by the Shandong Independent Innovation and Achievement Transformation Program (2014ZZCX07303), by the One Hundred-Talent Plan of Chinese Academy of Sciences (CAS) and by the Yantai Science and Technology Project (2013JH021)
Abstract: [Objective]Azorhizobium caulinodans ORS571 can fix nitrogen not only as a free-living organism and an associative-symbiotic bacterium by colonizing the root surface of non-leguminous plants, but also as a symbiotic bacterium by interacting with leguminous plant Sesbania rostrata. Due to its ability to grow and fix nitrogen under three conditions, A. caulinodans uses sophisticated chemotaxis signal transduction systems to transform environmental cues into corresponding behavioral responses. Chemotaxis appears crucial for the growth of A. caulinodansin complicated environment and the construction of associative relationship with the plant. However, little is known about the chemotactic pathway of A. caulinodans. Thus, our study aimed to compare the chemotaxis-like genes of A. caulinodans with those of well-studied species. [Methods]NCBI protein BLAST was used for searching sequence similarity with default parameter values against the genomes of A. caulinodans. HMMER3, based on Pfam database, was used for comparative analyses of methyl-accepting chemotaxis protein (MCP). [Results]There was a major chemotaxis cluster in A. caulinodans and the CheR methylated MCPs independently of pentapeptide motif. There were 43 MCP homologs containing diverse signal-sensing architectures in A. caulinodans. In addition, cytoplasmic domains of these MCPs were all composed of 38 heptad repeats. [Conclusion]Despite the extremely high homology presented between the chemotactic system of A. caulinodans and those of well-studied species, A. caulinodans shows its own unique characteristics. The classification of these chemotactic pathways by comparative genomics enables us to better understand how A. caulinodansresponds to changes in environment via exquisite signal transductions in chemotaxis system.
Key words: chemotaxis MCP Azorhizobium caulinodans
Azorhizobium caulinodans ORS571 is a microsymbiont isolated from the stem nodules of the tropical legume Sesbania rostrata, and has the capability of fixing nitrogen both under free-living and symbiotic conditions [1]. Nitrogen-fixing nodules are formed along the host stem as well as on the roots of S. rostrata [2]. Stem nodules arise at positions of adventitious root primordia after intercellular invasion by A. caulinodans [3].
Rhizobia move to the rhizosphere under the influence of chemotactic and growth-promoting compounds secreted by host plants, including amino acids, organic acids, sugars, aromatics, and various other secondary metabolites [4]. These chemical signals are exchanged via molecular dialogue between bacteria and plants. In Rhizobium leguminosarum bv. viciae, two chemotaxis gene clusters modulate cell motile behavior and swimming bias, thus benefit to nitrogen-fixing symbiosis within the roots of pea plants [5]. Chemotaxis is that bacteria move to the directed orientation relying on the sensitivity to chemical stimuli. A. caulinodans and other rhizobia such as Agrobacterium tumefaciens, Bradyrhizobium japonicum, and Rhizobium leguminosarum have the natural chemotactic traits to assemble among rhizosphere of their host [5-8].
Chemotaxis genes are widespread in a variety of microorganisms. The chemotaxis system of E. coli has been well studied and provides a paradigm for chemotaxis signal pathway. In E. coli, the chemotaxis pathway involves a major gene cluster containing most che genes, which are located closely to the flagella related genes [9]. Chemoreceptors, known as methyl-accepting chemotaxis proteins (MCPs), mediate gradient-tracking behavior via modulating CheA, a histidine kinase that acts as the central processing unit of the chemosensory circuit. CheA phosphorylates CheY and CheB, which are responsible for motor control and sensory adaptation respectively. The scaffold protein CheW couples with CheA to interact with receptors [10]. Phosphorylated CheY diffuses to the flagellar motor promoting a switch in the rotational direction from anticlockwise to clockwise [11]. CheY-P is hydrolyzed by phosphatase CheZ so that the promoting tumbling signal activity is extinguished. Methyltransferase CheR and methylesterase CheB mediate the reversible methylation of MCPs to stabilize the range of chemotactic sensitivity [10].
To date, little is known about chemotaxis system in Azorhizobium species even though whole genome sequence of A. caulinodans ORS571 had been annotated [12]. Comparative genome analysis of chemotaxis related genes of A. caulinodans with the publicly available che genes homologs in other species may be crucial for the understanding of their cellular functions and contributing to the study on their roles in symbiosis with host plant.
1 Materials and Methods 1.1 Multiple sequence alignment and phylogenetic analyses NCBI protein BLAST and position-specific-iterated-BLAST (blastp and psi-blast, respectively, http://blast.ncbi.nlm.nih.gov/) were used for searching sequence similarity with default parameter values against the genomes of A. caulinodans (GenBank: AP009384.1, http://www.ncbi.nlm.nih.gov/nuccore/AP009384.1)[13]. MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) was used with default values for parameter to conduct multiple sequence alignments and to establish the class membership of the methyl-accepting domains [14]. MEGA 4 software was used to construct 16S rRNA gene phylogenetic trees by the Neighbor-Joining [15].
1.2 Identification of chemotaxis related proteins and chemoreceptors from genomic data sets Chemotaxis related genes and proteins were retrieved from the genome of A. caulinodans ORS571. Chemoreceptors were identified by searching the NCBI nonredundant database for matches to the Pfam MCP signal domain profile (accession PF00015) with HMMER3[16]. Sequence logos were generated using WebLogo [17].
1.3 Promoter prediction and analysis Promoters were predicted with PromScan and Virtual Footprint using default parameters [18-19]. Only sequences fully match the typical promoter consensus sequence and lying within 300 bp upstream of predicted ORFs were considered.
2 Results and Discussion 2.1 General features of A. caulinodans ORS571 chemotaxis genes Scanning the whole genome of A. caulinodans, 50 genes of che and mcp had been annotated. It has a single chemotaxis pathway majorly located in one gene cluster containing cheA, cheW, cheY, cheB, and cheR (Azc 0661-0665), and the other two genes including cheY (Azc-0620) and cheZ (Azc-0621) are out of this cluster. All clusters are close to the flagellar gene clusters.
The che gene cluster in A. caulinodans was compared with those in the well-studied and genome-annotated α-proteobacterial species, several Rhizobiaceae were included. We constructed the phylogenetic tree using 16S rRNA genes, interestingly, the distribution and organization of che gene clusters appear consistent with the evolutionary relationship with 16S rRNA among these species (Figure 1). A. caulinodans owns the unique che gene order cheA, cheW, cheY, cheB, cheR (AWYBR), which was on the separate branch of the phylogenetic tree. The four species (A. tumefaciens, B. japonicum, R. leguminosarum and R. leguminosarum) on the top branch shared the che gene order YAWBRYD in the major functional cluster which controlled flagellar motility [20-21]. Clusters in B. japonicum and R. palustris were interrupted by several mcp genes between YAW and BR.
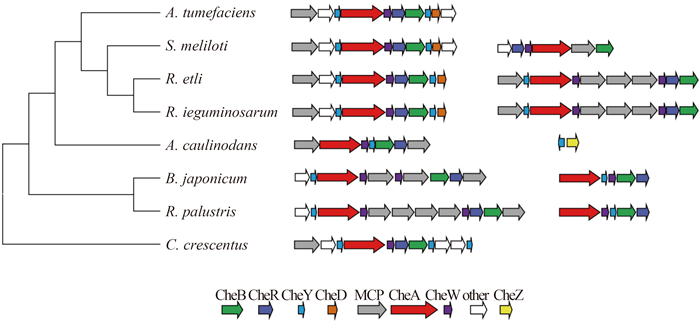 |
| Figure 1. Distribution and organization of che gene cluster in α-proteobacteria. One major cluster containing cheA is in A. caulinodans ORS571. The phylogenetic tree of the selected microorganisms is based on 16S rRNA sequences. |
| 图选项 |
A. caulinodans possessed a cheZ gene outside the chemotaxis gene neighborhood, which was indicated by the yellow arrow in Figure 1. CheZ is the well studied CheY-P phosphatase in E. coli [22] and rarely found in α-proteobacteria. CheZ can be distinguished by the conserved catalytic glutamine residue and high conservation of positions surrounding the catalytic residue in contrast with that in β/γ-proteobacteria [23]. A remote CheZ orthologue in Helicobacter pylori retains phosphatase function [24] and whether CheZ in A. caulinodans is activated awaits experimental analysis.
There is only one CheA homolog in A. caulinodans, but multiple cheA genes have been found in other species that thrive in natural environments [25]. In R. leguminosarum, genome encodes two chemotaxis gene clusters, namely che1 and che2, controlling swimming bias and chemotaxis. Nodulation and competition assays have demonstrated che1 is dominant pathway regulating cell motility and che2 has a minor effect on chemotaxis [8]. In A. tumefaciens and C. crescentus the che1 ortholog is determined to be the only chemotaxis operon controlling flagellar motility [20, 26]. We postulate that the only cluster containing CheA ortholog is the major pathways for chemotactic control and plant association in A. caulinodans. Previous experiments have proven our inference to some extent. A cheW mutant in A. caulinodans was previously found to be impaired in nodulation [27].
2.2 Distribution and characteristics of A. caulinodans MCPs 43 genes for MCPs have been discovered dispersing throughout A. caulinodans genome. These conjectural MCPs were identified by searching MA domains throughout the genome. The MA domain, being comprised of highly conserved amino acid sites, mediates biochemical signal when methylated by methyltransferase CheR [28]. Compared with five MCPs in E. coli [29], various MCPs plausibly reflect that A. caulinodans makes a physiological response that sense a vast range of environmental signals from external environment. E. coli owns five chemoreceptors that have similar structure with two transmembrane helices, HAMP and MA domains [30-31]. The sequences of the predicted A. caulinodans MCPs exhibit high diversity in the domain organization and topology (Figure 2-B).
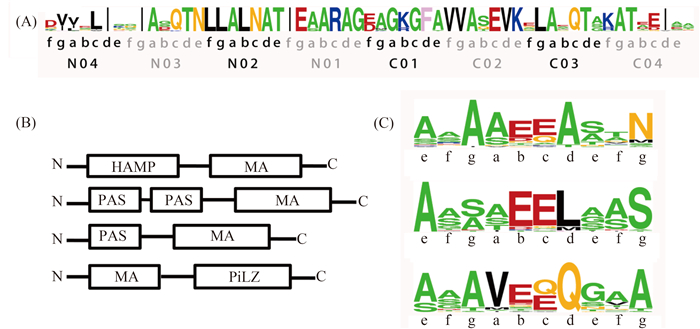 |
| Figure 2. Conserved subdomains and sites of MCPs in A. caulinodans. A: Class-specific conservation in the signaling subdomain. Eight heptads (N04-C04) in the centre of MCP-CD show extraordinarily high conservation. Each heptad is composed of intradimer (adeg) and interdimer (bcf) interaction sites. B: Domain architectures of MCPs in A. caulinodans show diversity. Abbreviation: HAMP, histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins and phosphatases; MA, methyl-accepting; PAS, the Drosophila period clock protein (PER), vertebrate aryl hydrocarbon receptor nuclear translocator (ARNT), and Drosophila single-minded protein (SIM); C: Methylation sites are conserved and located at specific positions. The potential methylation sites are the Glx pair is located in bc sites. Residue coloring: small (ASTG), green; hydrophobic (ILMV), black; aromatic (HFWY), yellow; negative (DE), red; polar (NQ), magenta; positive (KR), blue; and special (CP), cyan. |
| 图选项 |
In original studies on MCPs, the C-terminal cytoplasmic domains were recognized to be in a superfamily supported by multiple sequence alignment [32]. According to Zhulin’s research, the vast majority MCPs cytoplasmic domains (MCP-CD) in microbial chemoreceptors could be classified into seven category defined by the number of heptad repeats (namely 24H, 28H, 34H, 36H, 38H, 40H, 44H) [33]. By structural analysis, Tsr in E. coli and TM1143 in T. maritima belong to 36H and 44H, respectively [34-35]. Multiple sequence alignments of the A. caulinodans MCPs illustrated that all 43 MCPs belong to 38H, although some MCPs contain deletions in sequence.
Zhulin et al. have theorized that a MCP-CD consists of three subdomains. One signal domain in the central region and two methylation units on both sides are separated by two varied flexible bundle subdomains [33]. Eight Heptads in the centre of MCP-CD (N04-C04, Figure 2-A) show extraordinarily high conservation. All residues fall into intradimer (adeg) and interdimer (bcf) interaction sites, both of which contribute to bridging connections to CheA and CheW, but one stabilizes dimer by self-interaction and the other joints monomers by reverse linking [33]. Distinct residues still exist even though in highly conserved position such as site N03b of Tsr (36H-class) in E. coli, one Phe residue that is significant for receptor cooperativity [36-37]. In our studied 38H class, one Gln residue is conspicuous at C03c site, while it usually is one Arg residue in other classes. The function of this unique site on stabilizing trimer of dimmers relies on further experimental demonstrations.
To identify the potential methylation sites in MCPs, Glx pairs in the consensus methylation sequence -[ASTG]-[ASTG]-x(2)-[EQ]-[EQ]-x(2)-[ASTG]-[ASTG]- were searched in methylation subdomains [33]. Strikingly, two N-terminals and one C-terminal homologs were found in our 38H MCPs. The Glx pair is located in bc sites of heptads and small residues dock at upstream and downstream of Glx pair. This motif is thought to be critical for methyltransferase (CheR) and methylesterase (CheB) to implement reversible methylation in MCPs [38].
2.3 Characterization of the CheR tethering segment Sensory adaptation in bacterial chemotaxis is mediated by covalent modifications of specific glutamate and glutamine residues within the MCPs. In the well studied chemotaxis systems of E. coli, the high abundant receptors Tar and Tsr, sensing aspartic acid and serine in extracellular environment respectively, firm CheR through the interaction between the specific pentapeptide (NWETF) at the C-terminus and the β-subdomain of CheR [39]. In Thermotoga maritima, it is independent of C-terminal sequences for CheR to bind to MCPs, even though the last five residues show high conservation on hydrophobic residues at the third and fifth sites [40]. The two species mentioned above are representative of two typical CheR-pentapeptide-dependent and pentapeptide-independent respectively. The former has longer β-loop at β-subdomain. It appears that the three highly conserved glycine residues embedded in β-subdomain are important in enabling pentapeptide binding [41] (Figure 3). According to previous studies, the specific pentapeptide-containing MCPs belong to class 34H or to class 36H [33].
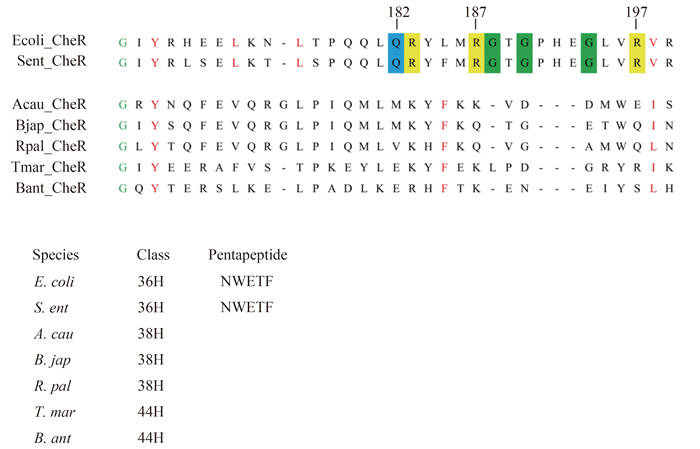 |
| Figure 3. Alignment of the β-subdomain of A. caulinodans CheR and other typical species. Conserved amino acid residues in β-subdomain are colored: small (G), green; hydrophobic (L, V, I, F, Y); amino acid residues that are conserved and that are proposed to be important for CheR-pentapeptide interactions are highlighted: small (G), green; positively charged (R, K), yellow; side-chain amine/amide containing residues (Q), blue. The organism abbreviation for each CheR homologue: Acau, Azorhizobium caulinodans; Bant, Bacillus anthracis; Bjap, Bradyrhizobium japonicum; Rpal, Rhodopseudomonas palustris; Sent, Salmonella enteric; Tmar, Thermotoga maritime. |
| 图选项 |
To identify which type A. caulinodans CheR belongs to, we compared the aligned β-subdomain sequences of A. caulinodans CheR with several representative species in different MCP classes. As shown in Figure 3, CheR homologues from species with 36Hs MCPs show high conservation at Gln182, Arg187, Arg197 and three Gly residues. These conserved residues have been proven to interact with specific residues in pentapeptide according to crystal structure analysis of S. enterica CheR [42]. A. caulinodans (38H) and T. maritime (44H) show less conservation except Gly166, Phe185, Ile/Leu198, which are highly conserved in both pentapeptide-dependent and independent β-subdomains. All three highly conserved residues are exposed to solvent and potentially available for interactions with other proteins [40]. Based on this alignment, we can deduce that the CheR in A. caulinodans may methylate MCPs independently of pentapeptide motif.
2.4 Prediction and analysis of che promter The bacterium has the capability to regulate the expression of its chemosensory machinery according to the external growth conditions [43]. The majority of genes in bacteria are transcribed by RNA polymerase bound to the specific factors σ70, σ28 and σ54. Both type of factors σ28 and σ54 widely participate in the transcription of che and flagellar gene clusters. The che and late flagellar gene such as lfgA to lfgN, are regulated by σ28 in E. coli [44]. In most cases, two sigma factors coordinate complex chemotaxis system collaboratively. In R. sphaeroides, one che cluster keeps basal transcriptional activity ensured by σ70 promoter and additional transcripts are initiated from the overlapping σ28, the other one involves in flagellar synthesis independently via σ54 [45].
We searched upstream of major A. caulinodans che operons for σ28 promoter consensus sequences, but no typical σ28 site was found. Then we searched for σ54 recognition site to determine whether it exhibits σ54 regulation. One classic -24/-12 consensus sequence was found upstream of the che cluster (Figure 4). Nonetheless, the predicted region needs to be further determined by electrophoretic mobility shift assay. In order to determine if A. caulinodans che gene expression is controlled in a hierarchic fashion with flagellar assembly, it will be necessary to analyze che expression in mutants that fail to synthesize polar and lateral flagella.
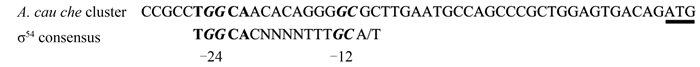 |
| Figure 4. Putative σ54 promoter elements. For the A. caulinodans major che cluster, the predicted transcription start site is 29 nucleotide bases upstream of predicted operon ATG start codons. –24/–12 consensus sequences are in italics. The initiation codon (underlined) is also indicated. |
| 图选项 |
In conclusion, the comparative analysis of chemotaxis related genes in A. caulinodans studied here has greatly refined our understanding of the function of the chemosensory pathways in A. caulinodans. The characteristic chemotaxis system may show superiority when rhizobia infect with nitrogen-desired roots of host plant. σ54- dependent regulation also reflects the complex chemotaxis mechanism to some extent.
External signals inputted into the chemosensory pathway from wide dynamic range tend to be diverse due to the presence of abundant chemoreceptor (mcp) genes. To date, there are no systematic studies on characterizing 38H MCPs-containing model bacterial strains, so it highlights the potential of A. caulinodans as a candidate of model organism for chemotaxis study. To make additional progress on the function of MCPs in signal transduction, deeper molecular insights at each organizational level are needed. The pursuit for the correct interplay between endosymbiosis and chemotaxis will propel the research of chemosensory pathway at the forefront of molecular study on biological signaling.
参考文献
| [1] | Donald RG, Ludwig RA. Rhizobium sp. strain ORS571 ammonium assimilation and nitrogen fixation.Journal of bacteriology, 1984, 158(3): 1144–1151. |
| [2] | Tsien HC, Dreyfus BL, Schmidt EL. Initial stages in the morphogenesis of nitrogen-fixing stem nodules of Sesbania rostrata.Journal of Bacteriology, 1983, 156(2): 888–897. |
| [3] | Goormachtig S, Capoen W, James EK, Holsters M. Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation.Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(16): 6303–6308. |
| [4] | Brencic A, Winans SC. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria.Microbiology and Molecular Biology Reviews, 2005, 69(1): 155–194. |
| [5] | Miller LD, Yost CK, Hynes MF, Alexandre G. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation.Molecular Microbiology, 2007, 63(2): 348–362. |
| [6] | Gulash M, Ames P, Larosiliere RC, Bergman K. Rhizobia are attracted to localized sites on legume roots.Applied and Environmental Microbiology, 1984, 48(1): 149–152. |
| [7] | Hawes MC, Smith LY. Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil-grown pea plants.Journal of Bacteriology, 1989, 171(10): 5668–5671. |
| [8] | Althabegoiti MJ, López-García SL, Piccinetti C, Mongiardini EJ, Pérez-Giménez J, Quelas JI, Perticari A, Lodeiro AR. Strain selection for improvement of Bradyrhizobium japonicum competitiveness for nodulation of soybean.FEMS Microbiology Letters, 2008, 282(1): 115–123. |
| [9] | Blattner FR, Plunkett Ⅲ G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12.Science, 1997, 277(5331): 1453–1462. |
| [10] | Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling.Current Opinion in Microbiology, 2010, 13(2): 124–132. |
| [11] | Dyer CM, Vartanian AS, Zhou HJ, Dahlquist FW. A molecular mechanism of bacterial flagellar motor switching.Journal of Molecular Biology, 2009, 388(1): 71–84. |
| [12] | Lee KB, De Backer P, Aono T, Liu CT, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, Najar FZ, Wiley GB, Roe B, Binnewies TT, Ussery DW, D'Haeze W, Herder JD, Gevers D, Vereecke D, Holsters M, Oyaizu H. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571.BMC Genomics, 2008, 9(1): 271. |
| [13] | Altschul SF, Madden TL, Sch?ffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucleic Acids Research, 1997, 25(17): 3389–3402. |
| [14] | Edgar R C. MUSCLE: multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Research, 2004, 32(5): 1792–1797. |
| [15] | Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0.Molecular Biology and Evolution, 2007, 24(8): 1596–1599. |
| [16] | Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching.Nucleic Acids Research, 2011, 39(Web Server issue): W29–W37. |
| [17] | Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator.Genome Research, 2004, 14(6): 1188–1190. |
| [18] | Studholme DJ, Buck M, Nixon T. Identification of potential sigma(N)-dependent promoters in bacterial genomes.Microbiology, 2000, 146 Pt 12: 3021–3023. |
| [19] | Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes.Bioinformatics, 2005, 21(22): 4187–4189. |
| [20] | Wright EL, Deakin WJ, Shaw CH. A chemotaxis cluster from Agrobacterium tumefaciens.Gene, 1998, 220(1/2): 83–89. |
| [21] | Greek M, Platzer J, Sourjik V, Schmitt R. Analysis of a chemotaxis operon in Rhizobium meliloti.Molecular Microbiology, 1995, 15(6): 989–1000. |
| [22] | Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.Nature Structural Biology, 2002, 9(8): 570–575. |
| [23] | Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis.Methods in Enzymology, 2007, 422: 1–31. |
| [24] | Lertsethtakarn P, Ottemann KM. A remote CheZ orthologue retains phosphatase function.Molecular Microbiology, 2010, 77(1): 225–235. |
| [25] | Tran HT, Krushkal J, Antommattei FM, Lovley DR, Weis RM. Comparative genomics of geobacter chemotaxis genes reveals diverse signaling function.BMC Genomics, 2008, 9: 471. |
| [26] | Alley MR, Gomes SL, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus.Genetics, 1991, 129(2): 333–341. |
| [27] | Suzuki S, Aono T, Lee KB, Suzuki T, Liu CT, Miwa H, Wakao S, Iki T, Oyaizu H. Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis.Applied and Environmental Microbiology, 2007, 73(20): 6650–6659. |
| [28] | Kort EN, Goy MF, Larsen SH, Adler J. Methylation of a membrane protein involved in bacterial chemotaxis.Proceedings of the National Academy of Sciences of the United States of America, 1975, 72(10): 3939–3943. |
| [29] | Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis.Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(1): 123–127. |
| [30] | Ma QH, Roy F, Herrmann S, Taylor BL, Johnson MS. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and flavin adenine dinucleotide binding.Journal of Bacteriology, 2004, 186(21): 7456–7459. |
| [31] | Watts KJ, Ma QH, Johnson MS, Taylor BL. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer.Journal of Bacteriology, 2004, 186(21): 7440–7449. |
| [32] | Le Moual H, Koshland DJ Jr. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis.Journal of Molecular Biology, 1996, 261(4): 568–585. |
| [33] | Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors.Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(8): 2885–2890. |
| [34] | Kristich CJ, Ordal GW. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis.Journal of Biological Chemistry, 2002, 277(28): 25356–25362. |
| [35] | Zimmer MA, Tiu J, Collins MA, Ordal GW. Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation.Journal of Biological Chemistry, 2000, 275(32): 24264–24272. |
| [36] | Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli.Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(10): 7060–7065. |
| [37] | Kim SH, Wang WR, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity.Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(18): 11611–11615. |
| [38] | Perez E, West AH, Stock AM, Djordjevic S. Discrimination between different methylation states of chemotaxis receptor Tar by receptor methyltransferase CheR.Biochemistry, 2004, 43(4): 953–961. |
| [39] | Wu JR, Li JY, Li GY, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation.Biochemistry, 1996, 35(15): 4984–4993. |
| [40] | Perez E, Stock AM. Characterization of the Thermotoga maritima chemotaxis methylation system that lacks pentapeptide-dependent methyltransferase CheR: MCP tethering.Molecular Microbiology, 2007, 63(2): 363–378. |
| [41] | Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase.Journal of Biological Chemistry, 2002, 277(44): 42325–42333. |
| [42] | Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine.Structure, 1997, 5(4): 545–558. |
| [43] | Shah DSH, Porter SL, Martin AC, Hamblin PA, Armitage JP. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes.The EMBO Journal, 2000, 19(17): 4601–4613. |
| [44] | Chevance FFV, Hughes KT. Coordinating assembly of a bacterial macromolecular machine.Nature Reviews Microbiology, 2008, 6(6): 455–465. |
| [45] | Martin AC, Gould M, Byles E, Roberts MAJ, Armitage JP. Two chemosensory operons of Rhodobacter sphaeroides are regulated independently by sigma 28 and sigma 54.Journal of Bacteriology, 2006, 188(22): 7932–7940. |
