赵峰1, 孟松松1, 2, 周德庆1


1. 中国水产科学研究院黄海水产研究所, 山东 青岛 266071;
2. 上海海洋大学食品学院, 上海 201306
收稿日期: 2015-06-08; 修回日期: 2015-09-10; 网络出版日期: 2015-09-14
基金项目: 国家自然科学基金(31201372)
通讯作者: 周德庆, Tel/Fax:+86-532-85819337;E-mail:zhoudq@ysfri.ac.cn
摘要: [目的] 构建副溶血弧菌庚糖基转移酶Ⅱ基因(waaF)的缺失株,探究waaF基因在副溶血弧菌O抗原合成中的作用。[方法] 本研究以副溶血弧菌临床分离株为研究对象,利用甲壳素介导的转化技术构建临床分离株的waaF基因缺失株;分别对野生株、缺失株的生长曲线、菌体形态和血清型进行了测定;利用大肠杆菌S17λpir菌株与副溶血弧菌结合转移的方法,分别构建O3、O5和O10来源的waaF基因的回补株,通过血清型测定,验证同源waaF基因的功能。[结果] 成功构建了waaF基因缺失株,基因缺失株生长正常,其生长曲线、菌体形态同野生菌株基本一致,基因缺失株同O抗血清不发生凝集反应,O抗原特性消失。回补实验显示,O3和O5来源waaF基因的回补株能恢复原有O抗原特性,O10来源waaF基因的回补株则不能恢复基因缺失株的O抗原特性。[结论] waaF基因同O抗原的合成相关,是O抗原合成的关键基因,不同O抗原副溶血弧菌中waaF基因功能存在差异。
关键词: 副溶血弧菌O抗原waaF基因功能分析
Gene deletion and functional analysis of the heptyl glycosyltransferaseⅡ (waaF) gene in Vibrio parahemolyticus O-antigen cluster
Feng Zhao1, Songsong Meng1, 2, Deqing Zhou1


1. Yellow Sea Fisheries Research Institute Chinese Academy of Fishery Sciences, Qingdao 266071, Shandong Province, China;
2. College of Food Science and Technology, Shanghai Ocean University, Shanghai 201306, China
Supported by the National Natural Science Foundation of China (31201372)
Corresponding author. Deqing Zhou, Tel/Fax: +86-532-85819337; E-mail: zhoudq@ysfri.ac.cn
Received: 8 June 2015; Revised: 10 September 2015; Published online: 14 September 2015
Abstract:[Objective] To construct heptyl glycosyltransferase gene II (waaF) gene deletion mutant of Vibrio parahaemolyticus, and explore the function of the waaF gene in Vibrio parahaemolyticus. [Methods] The waaF gene deletion mutant was constructed by chitin-based transformation technology using clinical isolates, and then the growth rate, morphology and serotypes were identified. The different sources (O3, O5 and O10) waaF gene complementations were constructed through E. coli S17λpir strains conjugative transferring with Vibrio parahaemolyticus, and the function of the waaF gene was further verified by serotypes. [Results] The waaF gene deletion mutant strain was successfully constructed and it grew normally. The growth rate and morphology of mutant were similar with the wild type strains (WT), but the mutant could not occurred agglutination reaction with O antisera. The O3 and O5 sources waaF gene complementations occurred agglutination reaction with O antisera, but the O10 sources waaF gene complementations was not. [Conclusion] The waaF gene was related with O-antigen synthesis and it was the key gene of O-antigen synthesis pathway in Vibrio parahaemolyticus. The function of different sources waaF gene were not the same.
Key words: Vibrio parahemolyticusO-antigenwaaF genefunctional analysis
副溶血弧菌(Vibrio parahaemolyticus)是一种革兰氏阴性嗜盐菌,属于弧菌属,广泛分布于海洋和河口环境中[1]。人们通常由于食用了生的或者加热不彻底的海产品感染副溶血弧菌,进而引起肠胃炎,典型的临床现象多为急性痢疾和腹部疼痛,伴有腹泻、恶心、呕吐、发低烧、发冷、和水样大便[2, 3]。自1996年以来,在全球范围内引起食物中毒事件爆发的致病性副溶血弧菌主要集中在少数几个血清型(O3:K6、O4:K68和O1:KUT等)中,这几种血清型构成了副溶血弧菌的主要流行群,其中O3:K6型在致病性副溶血弧菌中占80%左右,该流行群具有非常高的感染和传播能力[4, 5, 6]。Chen等的研究显示,上述流行群可能是由同一祖先,通过抗原基因簇及其邻近序列的重组进化而来,但副溶血弧菌的血清型多样性的遗传基础以及血清型同致病性之间的关系等尚不明确[7]。
脂多糖(LPS)是革兰氏阴性细菌的特有结构,在多数革兰氏阴性病原菌的致病过程中起到重要作用[8, 9]。典型的LPS包括三个部分,分别为特异性多糖侧链、脂质A及核心低聚糖。副溶血弧菌缺乏特异性多糖侧链,核心低聚糖可以作为其O抗原。副溶血弧菌共有13种O抗原类型(O1-O13),O抗原合成相关基因通常在细菌染色体上成簇分布,成为O抗原基因簇。副溶血弧菌标准菌株RIMD2210633的完整基因组测序完成后,基因预测显示位于大染色体区域A (VP0190-0214)可能为O抗原基因簇所在区域。革兰氏阴性菌O抗原基因簇中通常含有3种基因:单糖合成基因、糖基转移基因和寡糖单位处理基因。负责核心低聚糖合成的基因簇为waa基因簇(也称rfa基因簇),该类基因簇在革兰氏阴性菌中比较普遍,大肠杆菌中负责LPS的核心低聚糖装配的为rfa基因簇,目前已都被鉴定。在其他菌属中负责核心寡糖的基因簇被命名为waa基因簇,如沙门氏菌和肺炎克雷伯氏菌等[10, 11, 12]。waaF基因为副溶血弧菌标准菌株(RIMD2210633)区域A中编码庚糖基转移酶Ⅱ的基因,是合成核心低聚糖的重要酶,其功能的缺失会导致核心低聚糖结构的变化,进而可能导致副溶血弧菌的O抗原结构发生变化。
本文采用甲壳素介导的转化技术对临床分离的O3:K6菌株进行waaF基因的敲除,并对waaF基因缺失株进行回补实验,研究waaF基因缺失株的血清型是否发生变化,分析验证waaF基因是否为O抗原合成的关键基因。
1 材料和方法 1.1 材料1.1.1 菌株和质粒: 本实验所用副溶血性弧菌为临床分离株VP0178 (tdh+/trh-),血清型为O3:K6;环境分离株VP0037,血清型为O5:K15;环境分离株VP0043,血清型为O10:K24; E. coli S17 λpir菌株,具有KpnⅠ和XbaⅠ限制性酶切位点;所用质粒为pKD3(含氯霉素抗性基因),表达载体为pBBR1MCS2 (含卡那霉素抗性基因),以上菌株和质粒都由本实验保存。
1.1.2 主要试剂: 反转录试剂盒(RR047A),SYBR Green 试剂盒和LA Taq DNA聚合酶均购于宝生物工程(大连)有限公司;限制性内切酶Kpn I和XbaI,购于美国NEB公司。副溶血弧菌血清分型试剂购于日本生研公司;实验所用蟹壳来自青岛南山市场的梭子蟹。
1.1.3 引物: 本文所用引物由生工生物工程(上海)有限公司合成,引物核酸序列如表 1所示:
表 1. 本文所用引物序列Table 1. The primer sequences used in this paper
| Primers | Sequences (5′→–3′) |
| Sequence underline is the complementary sequence of the adjacent segment. GGTACC is the restriction enzyme cutting site of Kpn I, and CGG is the protective bases. TCTAGA is the restriction enzyme cutting site of Xba I, and TGC is the protective bases. | |
| PKD3-F | ATAAATAATTCGAATTAACATGAGCGATTGTGTAGGCTGGAG |
| PKD3-R | GACTGGCAAGTTGAAAAATGATTAACGGCTGACATGGGAATTAG |
| waaf-S-1000-F | CCCTGTATTATTTTCCACAAACT |
| waaf-S-1000-R | CTCCAGCCTACACAATCGCTCATGTTAATTCGAATTATTTATAC |
| waaf-X-1000-F | TCATTTTTCAACTTGCCAGTCAG |
| waaf-X-1000-R | AATTCCCATGTCAGCCGTTAAATCCAAACCATTCCGTTCCCTGA |
| waaf-n-F | AATGTTGGTATTGGCACTCACGC |
| waaf-n-R | ATTGGATACACTGCCTCGCCCTA |
| TLH-F | AAAGCGGATTATGCAGAAGCACTG |
| TLH-R | GCTACTTTCTAGCATTTTCTCTGC |
| TDH-F | GTAAAGGTCTCTGACTTTTGGAC |
| TDH-R | TGGAATAGAACCTTCATCTTCACC |
| HB-S-Kpn I | CGGGGTACCTTAACATGAACTAGTCGCAATGG |
| HB-X-Xba I | TGCTCTAGAATGAAAAAAATTCTAATTATTGG |
| S2-F | GCTTCGGAATCGTTTTCCGGGAC |
| S2-R | CTGCCCTGAACCGACGACCGGGT |
表选项
1.2 waaF基因缺失菌株的构建1.2.1 氯霉素抗性基因和waaF基因上下游同源臂的获得: 以质粒pKD3为模板扩增氯霉素抗性基因,以VP0178菌株基因组为模板扩增waaF基因上下游同源臂,所用引物对分别为PKD3-F/PKD3-R,waaf-S-1000-F/waaf-S-1000-R,waaf-X-1000-F/ waaf-X-1000-R。
PCR反应体系(50 μL)为:1 μL模板,1 μL上游引物(10 μmol/L),1 μL下游引物(10 μmol/L),25 μL PrimeSTAR Max Premix (TaKaRa),22 μL灭菌ddH2O。PCR程序参数:98℃ 2 min;98℃ 10s,55℃ 5 s,72℃ 1 min,进行30个循环;72℃ 10 min。
PCR产物经琼脂糖凝胶电泳,在凝胶成像系统下拍照记录结果。采用琼脂糖凝胶回收试剂盒,将氯霉素抗性基因、waaF基因上游同源臂和waaF基因下游同源臂进行回收纯化。
1.2.2 融合PCR法获得用于同源重组的DNA片段: 采用李敏等改进的融合PCR技术[13]将waaF基因上游同源臂、氯霉素抗性基因和waaF基因下游同源臂融合为同源重组所需的DNA片段。经琼脂糖凝胶电泳,回收获得纯化的融合片段。融合片段连接到pMD18-T载体,转化到TOP10大肠杆菌感受态细胞,经抗性平板筛选和菌落 PCR鉴定,挑取鉴定正确的阳性克隆,送北京华大基因测序。
1.2.3 甲壳素(蟹壳)介导的转化技术: 参考刘卫今等的方法,做了少量修改制备感受态细胞,具体如下:取VP0178菌株在TCBS培养基平板上划线,37℃培养12-16 h,挑取单菌落接种于含3%盐碱性蛋白胨水(APW)中,37℃、150 r/min,振荡培养6 h;将上述菌液以1:100的比例接种于3%盐APW中,37℃、160 r/min振荡培养2-3 h至OD600≈0.5左右;取1 mL上述菌液于2 mL离心管中,5000 r/min离心5 min沉淀菌体;取2 mL灭菌的海水重悬沉淀,使OD600≈0.2,作为感受态细胞,-80℃冰箱保存备用[14]。
参考Meibom等建立的甲壳素介导的转化方法,根据菌株特性做了修改用于构建基因缺失菌株,具体如下:取2 mL菌悬液于12孔细胞培养板中,并向孔中加入一片1 cm2的灭菌蟹壳,30℃培养过夜;弃菌液,仅留蟹壳,加入2 mL新鲜的灭菌海水,将2-4 μg融合DNA片段加入孔中,30℃ 培养过夜;取出蟹壳,放入3 mL 0.9%的生理盐水中,涡旋振荡30 s释放蟹壳上的菌体;将上述菌液用0.9%的生理盐水进行10-1、10-2、10-3梯度稀释,分别取100 μL梯度稀释菌液涂布在含5μg/mL 氯霉素抗性的LB平板上,培养18-24 h至长出单菌落;进行菌落PCR鉴定,获得waaF基因缺失株VP0178△waaF[15]。
1.3 副溶血弧菌waaF基因缺失回补菌株的构建1.3.1 构建重组质粒: 分别以VP0178、VP0037和VP0043菌株为模板扩增获得waaF-O3、waaF-O5 和waaF-O10基因片段,利用限制性内切酶Kpn I 和XbaⅠ对载体pBBR1MCS2和3种目的基因片段分别进行双酶切,并连接、转化E. coli S17 λpir感受态细胞,挑取平板上的可能阳性菌落,接种于加有1 mL的LB液体培养基中,37℃、160 r/min 培养6 h左右,至菌液浑浊,取1 μL菌液,进行PCR验证。对PCR鉴定为阳性的菌株,送华大基因进行测序,测序引物对为HB-S-Kpn I / HB-X-Xba I。
1.3.2 接合转移法构建副溶血弧菌基因回补株: 将含有3种waaF基因的pBBR1MCS2质粒的E. coli S17λpir菌株和VP0178△waaF菌株分别接种在LB肉汤中,37℃、160 r/min培养至OD600值为0.5左右;4℃、4000 r/min离心5 min收集菌体,用150 μL 的LB肉汤将细菌重新悬浮;取0.22 μm 微孔滤膜约1 cm2大小,紧贴在LB平板上,向滤膜上分别点加30 μL含有3种waaF基因的pBBR1MCS2质粒的E. coli S17λpir和VP0178△waaF菌株菌液,待菌液被滤膜充分吸收后,30℃倒置培养6-8 h;用0.9%的生理盐水洗涤微孔滤膜上的菌膜,将洗下的菌悬液进行10倍梯度稀释,每个梯度取200μL涂布在LB平板(50μg/mL Kan+)上,30℃正置30 min,然后将平板倒置,30℃培养至长出单菌落;挑取平板上的单菌落,接种于加有1 mL LB液体培养基的2 mL离心管中,37℃、160r/min培养6 h左右,待菌液浑浊,取1 μL菌液,进行PCR验证[18]。
1.4 生长曲线的绘制取VP0178菌株和VP0178△waaF菌株在TCBS培养基平板上划线,37℃过夜培养。挑取单菌落接种于3%盐APW水中,37℃、160r/min 培养6 h,按4%接种量将活化的菌液接种于含3%盐APW水的摇瓶中,37℃、160 r/min培养菌体,每隔2 h测定1次OD600值,直到各个菌进入平台期,绘制生长曲线。
1.5 革兰氏染色及镜检取VP0178菌株和VP0178△waaF菌株在TCBS培养基平板上划线,37℃过夜培养。用0.9%的生理盐水冲洗平板上的菌落至冲洗液浑浊,作为待检样品进行革兰氏染色镜检。
1.6 血清型鉴定副溶血弧菌的O和K血清分型按照日本生研公司血清鉴定试剂盒说明书进行。
2 结果和分析 2.1 副溶血弧菌waaF基因缺失菌株VP0178△waaF的构建及鉴定分别以VP0178菌株和pKD3质粒为模板,扩增获得waaF基因上游同源臂(1230 bp)、下游同源臂(1143 bp)和氯霉素抗性基因片段(1053 bp),具体扩增结果如图 1-A中所示。将上述3个扩增片度回收后,采用融合PCR,将3个片段连接起来,融合PCR扩增结果如图 1-B所示,从图中可看出,通过融合PCR获得了片段大小约为3000 bp的条带,同预期片段大小3426 bp一致。融合PCR片段的测序结果显示,片段中依次包含了waaF基因上游同源臂、氯霉素抗性基因片段和waaF基因下游同源臂,同预期结果一致,表明融合PCR扩增成功。
 |
| 图 1. 各目的片段的琼脂糖凝胶电泳图 Figure 1. Agarose gel electrophoresis of the target fragments. A and B showed the results of construction of vector, M1: DL2000 maker, M2: DL5000 maker; lane 1: upstream homologous arm of waaF gene; lane 2: downstream homologous arm of waaF gene; lane 3: chloramphenicol resistant gene cloning from pKD3 plasmid; lane 4: fusion segment. C showed the result of identification of gene deletion strains; lane 5: fragments amplified by primers waaf-n-F and waaf-n-R; lane 6: fragments amplified by primers waaf-S-1000-F and waaf-X-1000-R; lane 7: fragments amplified by primers PKD3-F and PKD3-R; lane 8: fragments amplified by primers TLH-F and TLH-R; lane 9: fragments amplified by primers TDH-F and TDH-R. |
| 图选项 |
将融合片段经甲壳素介导的转化进入到VP0178菌种,经抗性标记筛选,获得初步阳性克隆,经菌落PCR鉴定,获得VP0178△waaF菌株。图 1-C中显示了PCR验证正确菌株的扩增结果,其中泳道5为waaF基因内部引物waaf-n-F和waaf-n-R扩增结果,因为waaF基因已敲除,故该引物无法获得扩增片段。泳道6为waaF基因上游同源臂正向引物和下游同源臂反向引物waaf-S-1000-F和waaf-X-1000-R的扩增结果,在VP0178△waaF菌株中,由于缺失了waaF基因,其大小应为3426 bp。泳道7为pKD3载体抗性基因引物PKD3-F和PKD3-R的扩增结果,在VP0178△waaF菌株中有插入的抗性基因,其大小应为1053 bp。8为副溶血弧菌特征性引物TLH-F和TLH-R的扩增结果,其大小为500 bp,该片段为副溶血弧菌特有。9为副溶血弧菌毒力基因引物TDH-F和TDH-R的扩增结果,基因缺失的野生菌株VP0178为临床分离株,tdh基因检测为阳性,片段大小为269 bp,在VP0178△waaF菌株也应含有该片段。上述片段的扩增结果同预期相一致,显示VP0178△waaF菌株构建成功。
2.2 VP0178△waaF的菌体形态将VP0178菌株和VP0178△waaF菌株分别点种于TCBS平板上,37℃培养18 h,观察菌体形态,结果显示两株菌均长成蓝绿色菌落,两株菌的形态较为接近,如图 2-A所示。经革兰氏染色,观察VP0178菌株和VP0178△waaF菌株的菌体形态,结果如图 2-B和图 2-C所示,VP0178菌株和VP0178△waaF菌株的菌体形态基本一致,革兰氏染色图均为短杆状,说明waaF基因的缺失没有影响细胞的形态。
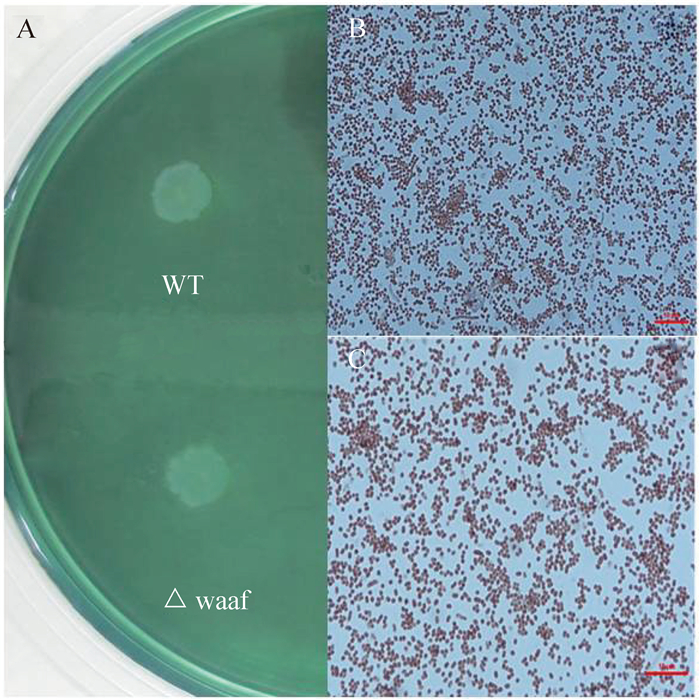 |
| 图 2. VP0178和VP0178△waaF的生长和菌体形态 Figure 2. The growth and gram staining of VP0178 and VP0178△waaF. A showed the growth of VP0178 and VP0178△waaF strain on TCBS agar, WT: VP0178 strain, △waaf: VP0178△waaF mutants strain; B and C showed the gram staining of VP0178 and VP0178△waaF strain, respectively. |
| 图选项 |
2.3 VP0178△waaF的生长曲线将VP0178菌株和VP0178△waaF菌株分别接种在3%盐的APW水中,同时测定VP0178菌株和VP0178△waaF菌株的生长速率,结果显示如图 3 中所示,VP0178菌株和VP0178△waaF菌株的生长速率基本一致,VP0178△waaF菌株生长正常,说明waaF基因的缺失没有影响细胞的生长。
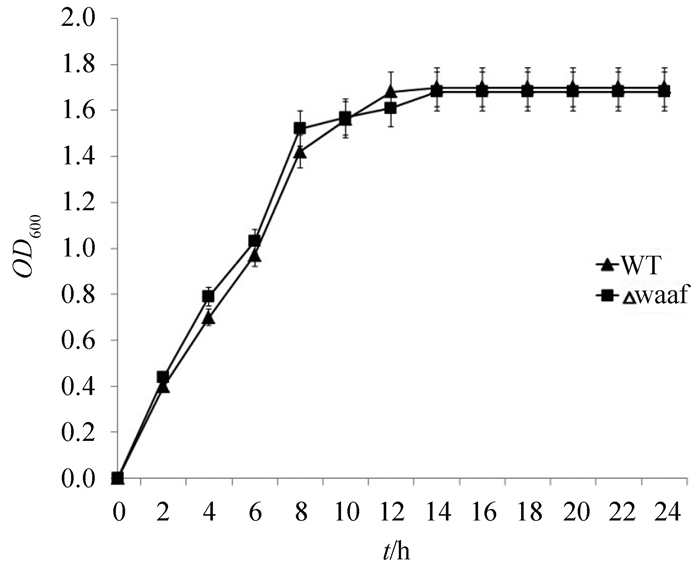 |
| 图 3. VP0178和VP0178△waaF的生长曲线 Figure 3. The growth rate and gram staining of VP0178 and VP0178△waaF. WT: VP0178 strain, △waaf: VP0178△waaF mutants strain |
| 图选项 |
2.4 VP0178△waaF的血清型鉴定使用O3抗血清分别同野生菌株(VP0178)和基因缺失菌株(VP0178△waaF)进行反应,结果如图 4中所示,野生型同O3抗血清发生了明显的凝集反应,显示为O3血清型;而VP0178△waaF菌株同O3抗血清没有发生凝集反应,显示VP0178△waaF菌株失去了O3血清表型。将VP0178△waaF菌株同O1、O2和O4-O11抗血清进行反应,结果显示VP0178△waaF菌株同这10种抗血清均不发生凝集反应。上述结果表明waaF基因在O抗原合成中具有重要作用,其缺失直接影响O抗原的合成。
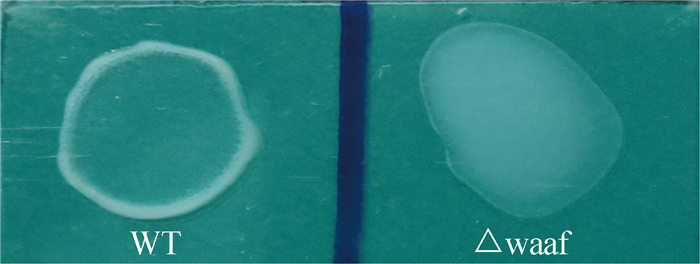 |
| 图 4. 野生菌株与基因缺失株的O3血清型鉴定对比图 Figure 4. Comparison of the reaction results with O3 antiserum between WT and mutant strains. WT: VP0178, △waaf: VP0178△waaF mutant. |
| 图选项 |
2.5 VP0178△waaF回补菌株的构建及功能分析为了进一步验证waaF基因的功能,构建了VP0178△waaF回补菌株。选取了野生菌株的waaF基因(O3来源),同野生菌株相似度较高的waaF基因(O5来源,相似度99.3%)和同野生菌株相似度较低的waaF基因(O10来源,相似度53.1%),利用pBBR1MCS2质粒分别构建了3个回补表达载体:pBBR1MCS2-waaf-O3、pBBR1MCS2-waaf-O5和 pBBR1MCS2-waaf-O10。图 5中显示的是3个载体构建成功的PCR验证结果,从图中可以看出使用pBBR1MCS2质粒上S2-F 和S2-R引物在空质粒上扩增出1006 bp大小片带(图 5中泳道1),在其余3个回补表达载体中分别扩增出2065 bp (1006 bp+1059 bp)、1732 bp (1006 bp+726 bp)和2065 bp (1006 bp+1059 bp)大小的片段,同理论值一致,表明3个重组表达载体构建成功。
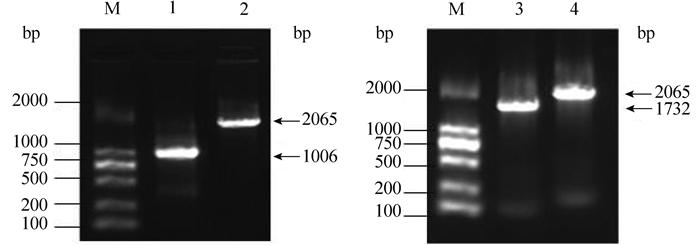 |
| 图 5. 回补表达载体的琼脂糖凝胶电泳图 Figure 5. Agarose gel electrophoresis of the PCR products of the recombinant plasmids. M: DL2000 maker; lane 1: empty plasmid; lane 2: pBBR1MCS2-waaF-O3; lane 3: pBBR1MCS2-waaF-O10; lane 4: pBBR1MCS2-waaF-O5. |
| 图选项 |
3个回补表达载体使用结合转移方法经E. coli S17λpir菌株转入到VP0178△waaF菌株中,经卡那霉素抗性平板筛选、PCR验证和测序验证,获得构建成功的VP0178△waaF回补菌株。图 6显示的3个回补菌株的血清型鉴定结果,其中含有野生株waaF基因的回补株及含有pBBR1MCS2-waaF-O5质粒的回补株同O3抗血清发生了凝集反应,说明野生株waaF基因的回补使得缺失株恢复了O3血清型,O5血清型的waaF基因能在O3血清型的副溶血弧菌株中发挥庚糖基转移酶功能;而含有空质粒pBBR1MCS2及pBBR1MCS2-waaF-O10质粒的回补株同O3抗血清不发生凝集反应,说明pBBR1MCS2质粒及O10血清型的waaF基因对O3表型不起作用。
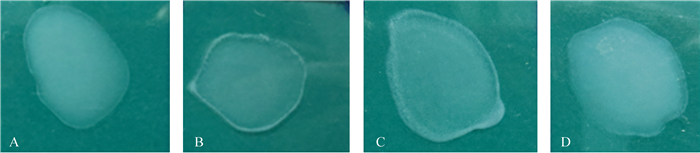 |
| 图 6. 3个基因回补株的O3血清型鉴定对比图 Figure 6. Comparison of the reaction results with O3 antiserum among the gene complementations. A: C-0, mutant complemented by empty plasmid pBBR1MCS2; B: C-△waaF-O3, mutant complemented by pBBR1MCS2-waaF-O3; C: C-△waaF-O5, mutant complemented by pBBR1MCS2-waaF-O5; D: C-△waaF-O10, mutant complemented by pBBR1MCS2-waaF-O10. |
| 图选项 |
3 讨论在细菌基因的敲除过程中,将外源基因高效率地转化到靶细胞中是基因敲除过程中至关重要的一步,然而包括副溶血弧菌在内的野生型弧菌,因其产生胞外DNase而很难实现外源基因的高效率转化[16, 17]。传统的化学转化和电转化法很难能够实现弧菌的成功转化。Hamashima和其同事已经将质粒pBBR322,pACYC18和 pHSG398通过电转化成功导入副溶血弧菌的感受态细胞,但是转化效率低,仅能转化102个转化体/μg DNA[18]。Kawagishi和其同事研究发现在同副溶血弧菌亲缘关系较近的溶藻弧菌中,电转化前去除细胞外DNase能够提高转化效率[19],然而这种方法却不适用于副溶血弧菌,因为副溶血弧菌对去除细胞外DNase所需要营造的低渗透压环境非常敏感。
Meibom等研究发现,弧菌属的创伤弧菌、霍乱弧菌和副溶血弧菌等存在自然重组现象,即生长在蟹壳等甲壳素表面的弧菌能够从环境中获得游离的DNA,可将其整合到自己的基因组中,通过天然的竞争自然重组,它们可以更好的适应环境或更容易使人类致病[15]。根据这一自然重组的原理,可利用蟹壳等甲壳素可建立外源基因导入到弧菌中的重组技术,Gulig等报道了创伤弧菌甲壳素介导的同源重组技术。本文以副溶血弧菌为实验菌株,利用甲壳素介导的基因敲除技术成功的敲除waaF基因,该技术所需实验条件简单,操作步骤便捷,基因敲除效率较高,这为后续开展副溶血弧菌中功能基因的验证建立了可靠的方法[20]。
O抗原存在于细胞表面,是主要的表面抗原,在进化过程中承受着较大的选择压力,因而变异速度也较快,呈现出显著的遗传多样性。对O抗原相关基因研究比较深入的是小肠杆菌、沙门氏菌、霍乱弧菌等,有关副溶血弧菌O抗原的合成途径和合成关键基因的研究较少。本文通过构建waaF基因缺失株,进行血清型验证,结合同源waaF基因的回补实验,证实waaF基因同O抗原的合成有关,其缺失会导致O抗原表型丧失,不同O抗原副溶血弧菌中waaF基因的功能存在差异。为探索副溶血弧菌的O抗原的合成途径、分子进化机制等提供技术手段。
副溶血弧菌致病的毒力因子包括:黏附因子、侵袭因子、溶血毒素、III型分泌系统等,其中黏附因子是病原菌接触和感染宿主的第一步,也是引起感染,导致宿主疾病的首要条件[21]。黏附作用主要是通过病原菌表面的特殊结构和表面物质与宿主细胞表面受体结合并发生黏附作用,但目前对副溶血弧菌如何定殖与人或其他宿主体内的机制还不十分明确,在多种革兰氏阴性细菌中,O抗原都与细菌同宿主的黏附作用相关[22]。本文构建了O抗原合成基因——waaF基因缺失的突变株,可为下一步探索O抗原同副溶血弧菌粘附力的关系、O抗原同副溶血弧菌致病力之间的关系提供条件。
参考文献
| [1] | Bej AK, Patterson DP, Brasher CW, Vickery MCL, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. Journal of Microbiological Methods, 1999, 36(3): 215–225. |
| [2] | Shimohata T, Takahashi A. Diarrhea induced by infection of Vibrio parahaemolyticus. The Journal of Medical Investigation, 2010, 57(3/4): 179–182. |
| [3] | Yeung PSM, Boor KJ. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathogens & Disease, 2004, 1(2): 74–88. |
| [4] | Chowdhury A, Ishibashi M, Thiem VD, Tuyet DTN, Van Tung T, Chien BT, von Seidlein L, Canh DG, Clemens J, Trach DD, Mishibuchi M. Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiology and Immunology, 2004, 48(4): 319–327. |
| [5] | Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3: K6 and its serovariants. Clinical Microbiology Reviews, 2007, 20(1): 39–48. |
| [6] | Velazquez-Roman J, León-Sicairos N, Flores-Villase?or H, Villafa?a-Rauda S, Canizalez-Roman A. Association of pandemic Vibrio parahaemolyticus O3: K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Applied and Environmental Microbiology, 2012, 78(6): 1794–1803. |
| [7] | Chen YS, Stine OC, Badger JH, Gil AI, Nair GB, Nishibuchi M, Fouts DE. Comparative genomic analysis of Vibrio parahaemolyticus: serotype conversion and virulence. BMC Genomics, 2011, 12(1): 294. |
| [8] | Erwin AL, Allen S, Ho DK, Bonthius PJ, Jarisch J, Nelson KL, Tsao DL, Unrath WCT, Watson ME Jr, Gibson BW, Apicella MA, Smith AL. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infection and Immunity, 2006, 74(11): 6226–6235. |
| [9] | Ho DK, Ram S, Nelson KL, Bonthuis PJ, Smith AL. lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. The Journal of Immunology, 2007, 178(2): 1002–1012. |
| [10] | Kanipes MI, Holder LC, Corcoran AT, Moran AP, Guerry P. A deep-rough mutant of Campylobacter jejuni 81–176 is noninvasive for intestinal epithelial cells. Infection and Immunity, 2004, 72(4): 2452–2455. |
| [11] | Swords WE, Buscher BA, Ver Steeg Ii K, Preston A, Nichols WA, Weiser JN, Gibson BW, Apicella MA. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Molecular Microbiology, 2000, 37(1): 13–27. |
| [12] | Chen M, Guo D, Wong HC, Zhang X, Liu FX, Chen HY, Chen M, Liu B, Wang L, Wu F, Feng L. Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. International Journal of Food Microbiology, 2012, 159(2): 122–129. |
| [13] | Li M, Yang Q. A Rapid method for generation of homologous recombinant fragments-fusion PCR. China Biotechnology, 2007, 27(8): 53–58. (in Chinese). 李敏, 杨谦. 一种高效构建同源重组DNA片段的方法——融合PCR. 中国生物工程杂志, 2007, 27(8): 53–58. |
| [14] | Liu WJ, Jiang Y, Wan DD. Study on a simple and efficient preparation of competent cell and the establishment of plasmid transformation system. Journal of Anhui Agricultural Sciences, 2008, 36(7): 2745–2746. (in Chinese) 刘卫今, 蒋勇, 完迪迪. 简单高效的感受态细胞制备和质粒转化体系的建立. 安徽农业科学, 2008, 36(7): 2745–2746. |
| [15] | Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science, 2005, 310(5755): 1824–1827. |
| [16] | Newland JW, Green, BA, Foulds J, Holmes RK. Cloning of extracellular DNase and construction of a DNase-negative strain of Vibrio cholerae. Infection and Immunity, 1985, 47(3): 691–696. |
| [17] | Focareta T, Manning PA. Distinguishing between the extracellular DNases of Vibrio cholerae and development of a transformation system. Molecular Microbiology, 1991, 5(10): 2547–2555. |
| [18] | Hamashima H, Nakano T, Tamura S, Arai T. Genetic transformation of Vibrio parahaemolyticus, Vibrio alginolyticus, and Vibrio cholerae Non O-1 with Plasmid DNA by Electroporation. Microbiology and Immunology, 1990, 34(8): 703–708. |
| [19] | Liu X, Gao H, Yang L, Zhang YQ, Tan YF, Guo ZB, Huang XX, Yang RF, Zhou DS. Establishment of a suicide vector-based gene knockout method in studies of Vibrioparahaemolyticus. Acta Laboratorium Animalis Scientia Sinica, 2011, 19(3): 188–192. (in Chineses) 刘霞, 高鹤, 杨琳, 张义全, 谭亚芳, 郭兆彪, 黄新祥, 杨瑞馥, 周冬生. 副溶血性弧菌基因敲除方法的建立及应用. 中国实验动物学报, 2011, 19(3): 188–192. |
| [20] | Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Applied and Environmental Microbiology, 2009, 75(15): 4936–4949. |
| [21] | Whitaker WB, Richards GP, Boyd EF, Payne SM. Loss of sigma factor RpoN increases intestinal colonization of Vibrio parahaemolyticus in an adult mouse model. Infection and Immunity, 2014, 82(2): 544–556 |
| [22] | Chen YS, Bystricky P, Adeyeye J, Panigrahi P, Ali A, Johnson JA, Bush CA, Morris JG, Stine OC. The capsule polysaccharide structure and biogenesis for non-O1 Vibrio cholerae NRT36S: genes are embedded in the LPS region. BMC Microbiology, 2007, 7(1): 20. |
