 , 唐荧霜2, 丁阿强1,2
, 唐荧霜2, 丁阿强1,2
 , 王学文2, 李微薇2, 柴风光2, 张代钧1,2
, 王学文2, 李微薇2, 柴风光2, 张代钧1,21. 重庆大学煤矿灾害动力学与控制国家重点实验室, 重庆 400044;
2. 重庆大学环境科学系, 重庆 400044
收稿日期: 2018-11-12; 修回日期: 2019-01-12; 录用日期: 2019-01-12
基金项目: 中央高校基本科研业务费专项资金(No.2018CDXYZH0006,2019CDQYZH002);重庆市基础科学与前沿技术研究专项项目(No.CSTC2016jcyjA0506,CSTC2018jcyjAX0802)
作者简介: 卢培利(1975-), 男, E-mail:lupl@cqu.edu.cn
通讯作者(责任作者): 丁阿强, E-mail:aqding@cqu.edu.cn
摘要: 亚硝酸盐型厌氧甲烷氧化(Nitrite-dependent anaerobic methane oxidation,n-DAMO)是微生物在厌氧条件下利用甲烷还原亚硝酸盐的过程.本研究通过排泥的策略对n-DAMO过程进行强化,并比较分析了反应器中微生物的群落结构及功能微生物数量.结果发现,与对照组相比,排泥后实验组反应器的脱氮速率从17.00 mg·L-1·d-1提高到73.10 mg·L-1·d-1.排泥后反应器中n-DAMO细菌的相对丰度从38.3%上升到67.7%,功能微生物的基因拷贝数由1.404×108 copies·g-1增长到4.854×108 copies·g-1,污泥比活性提高了2.95倍.与之相反,初始反应器中其余优势微生物Unclassified_GCA004、Unclassified_Rhodocyclaceae、Unclassified_Fimbriimonadaceae与Methylosinu相对丰度分别下降为原来的27.66%、32.65%、4.35%、20.27%.结果表明,排泥可以有效地强化n-DAMO过程,同时促进功能微生物的生长,主要原因在于排泥排出了非目标微生物,使得目标微生物大量生长.本研究为强化n-DAMO过程及加快n-DAMO微生物的富集提供了一条新思路,并为进一步推动n-DAMO过程的工程应用提供了理论基础.
关键词:亚硝酸盐型厌氧甲烷氧化排泥脱氮效能微生物群落结构生长速率
New insight into the enhancement of nitrite-dependent anaerobic methane oxidation: Withdrawing sludge and its microbial mechanism
LU Peili1,2
 , TANG Yingshuang2, DING Aqiang1,2
, TANG Yingshuang2, DING Aqiang1,2
 , WANG Xuewen2, LI Weiwei2, CHAI Fengguang2, ZHANG Daijun1,2
, WANG Xuewen2, LI Weiwei2, CHAI Fengguang2, ZHANG Daijun1,2 1. State Key Laboratory of Coal Mine Disaster Dynamics and Control, Chongqing University, Chongqing 400044;
2. Department of Environmental Science, Chongqing University, Chongqing 400044
Received 12 November 2018; received in revised from 12 January 2019; accepted 12 January 2019
Abstract: Specific microorganisms involving in nitrite-dependent anaerobic methane oxidation (n-DAMO) process could utilize methane to reduce nitrite under anaerobic condition. In this study, the effect of withdrawing sludge on n-DAMO process was investigated and its microorganism community as well as functional microorganism abundance were analyzed. Compared with control group, the nitrogen removal rate of experimental group increased from 17.00 mg·L-1·d-1 to 73.10 mg·L-1·d-1 significantly after withdrawing sludge. Microorganism community analysis showed that the relative abundance of n-DAMO bacteria increased from 38.3% to 67.7% after withdrawing sludge. Functional microorganism abundance increased from 1.404×108 copies·g-1 to 4.854×108 copies·g-1 and sludge specific activity increased by 295%. In contrast, the relative abundances of unclassified_GCA004, unclassified_Rhodocyclacea, unclassified_Fimbriimonadaceae and Methylosinu decreased to 27.66%, 32.65%, 4.35% and 20.27%, respectively, after withdrawing sludge. The results indicate that withdrawing sludge can strengthen n-DAMO process and promote functional microorganism growth, which might be due to the increase of n-DAMO functional microorganisms abundance in the system. This study gives a new insight to strengthen n-DAMO process and accelerate n-DAMO microorganism enrichment, which provides theoretical basis for the application of n-DAMO process.
Keywords: nitrite-dependent denitrifying anaerobic methane oxidationwithdrawing sludgenitrogen removal efficiencymicrobial community structuregrowth rate
1 引言(Introduction)近年来, 由于工农业废水的排放, 导致我国地表水体氮素含量不断升高, “水华”、“赤潮”现象频发, 对农业、渔业、旅游业等造成了巨大影响.目前, 硝化-反硝化生物脱氮工艺普遍面临有机碳源不足、反硝化不充分、总氮达标难度大的困境, 而额外投加碳源不仅大大增加了废水脱氮成本, 且会进一步引起温室气体甲烷的大量排放(Solomon et al., 2007; 沈李东等, 2011), 加剧温室效应.鉴于此, 低碳耗脱氮技术开发或者经济碳源替代成为可能的解决途径.
硝态氮型厌氧甲烷氧化(NOx-Dependent Anaerobic Methane Oxidation, NOx-DAMO)是一种新兴的生物脱氮过程, 其以硝酸盐或亚硝酸盐为电子受体, 以甲烷为电子供体, 可以同步脱除氮素污染并缓解温室效应.NOx-DAMO过程的反应式如下所示:
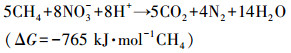 | (1) |
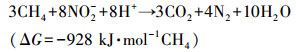 | (2) |
由于n-DAMO微生物增殖缓慢, 富集量少, 因此, 研究者都在尽量保存生物量, 极少研究排泥对n-DMAO活性的影响.但培养体系中存在少量其他竞争微生物和抑制性产物, 均会对n-DAMO微生物的生长产生干扰, 故可以通过排泥的方式对n-DAMO微生物进行加速富集并提高活性.基于此, 本研究以实验室稳定运行的序列式活性污泥法反应器(SBR)中n-DAMO功能微生物为研究对象, 考察排泥对n-DAMO微生物及脱氮过程的影响, 并利用荧光定量PCR技术、高通量测序技术分析功能微生物与其余微生物群落结构变化, 进一步解析内在的微生物学机制, 以期进一步丰富n-DAMO过程的强化策略.
2 材料与方法(Materials and methods)2.1 反应器构型实验所采用的反应装置如图 1所示(柴风光等, 2018; Li et al., 2018), 反应器材质为双层玻璃, 有效容积为2 L, 反应器内液相体积占1.5 L, 气相体积占0.5 L, 外隔层为水浴区.反应器内置搅拌子, 使用磁力搅拌器进行搅拌.
图 1(Fig. 1)
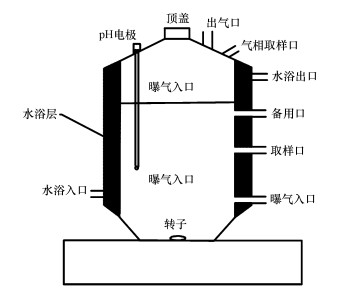 |
| 图 1 SBR示意图 Fig. 1Diagram of sequence batch reactor |
2.2 接种污泥与培养液本研究以人工景观湖底泥(60 mL)、城市污水处理厂污泥浓缩池厌氧污泥(400 mL)、稻田土沉积物(取表层泥以下10 cm处污泥450 mL)混合物为接种物.接种污泥沉淀并弃去上清液, 再混合均匀过10目筛, 在连续曝氮气条件下用纯水冲洗污泥3~5次.最后取380 mL装入SBR反应器中, 并加入1.5 L培养液(柴风光等, 2018; Li et al., 2018), 成分如表 1所示(Ettwig et al., 2009).矿物基质、酸性微量元素、碱性微量元素分开配制, 每升基质中添加0.5 mL酸性微量元素(包含0.1 mol · L-1 HCl)、0.2 mL碱性微量元素(包含0.1 mol · L-1 NaOH).
表 1(Table 1)
| 表 1 微量元素与矿物基质表 Table 1 Trace elements and mineral matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
表 1 微量元素与矿物基质表 Table 1 Trace elements and mineral matrix
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
2.3 反应器运行设置两组相同的SBR反应器, 分别在严格厌氧条件下运行, 其中, 实验组记为1号, 对照组记为2号.维持反应器中亚硝酸盐浓度为30 mg · L-1, 水力停留时间为90 d, 反应温度为30 ℃, pH通过1 mol · L-1的HCl维持在7.0~7.5之间.1号反应器在实验第42 d排泥500 mL, 排泥比为33.3%;在第172 d排泥250 mL, 排泥比为16.7%.实验期间两只反应器每天曝气10 min(体积分数占比:5%N2、90%CH4、5%CO2)(Lu et al., 2019), 曝气速率为150 mL · min-1, 反应器内压力维持在1.01×105~1.11×105 Pa.
2.4 分析方法2.4.1 化学分析水样经0.45 μm滤膜过滤后, 通过N-(1-萘基)-乙二胺光度法测定亚硝酸盐浓度.脱氮速率通过线性拟合进行计算, 具体方法为反应前4 h每一小时取样测定亚硝酸盐浓度, 以时间为横轴、以亚硝氮为纵轴拟合直线, 斜率即为最大脱氮速率.通过气相色谱法(SC-3000B, 重庆川仪自动化股份有限公司)测定氮气、甲烷浓度, 并利用理想气体方程和亨利公式计算反应器内总氮气量和总甲烷量.
2.4.2 高通量测序分析取第132 d和第214 d对照组与实验组反应器中污泥作为样本, 利用DNA提取试剂盒抽提样本基因组DNA, 选用细菌通用引物338F(5′-ACTCCTACGGGAGGCAGCAG-3′)和806R(5′-GGACTACHVGGGTWTCTAAT-3′)进行PCR扩增.PCR产物经纯化与定量后上机(Illumina Miseq PE250/PE300平台上海派森诺生物科技有限公司).
2.4.3 实时荧光定量PCR分析以提取的基因组DNA为模板, 选用qP1F (GGGCTTGACATCCCA CGAACCTG)和qP1R (CGCCTTCCTCCAGCTTG ACGC)(Ettwig et al., 2009)引物进行定量扩增.扩增程序为:95 ℃预变性3 min, 95 ℃变性15 s, 60 ℃退火30 s, 72 ℃延伸30 s, 进行40个循环, 72 ℃再延伸5 min.每个样品均有3个生物学重复, 基因拷贝数以污泥干重计.
3 结果与讨论(Results and discussion)3.1 排泥对n-DAMO反应器脱氮效能影响两个SBR反应器的亚硝酸盐浓度如图 2所示.对每天前4 h的亚硝酸盐浓度-时间曲线进行拟合, 发现符合零级动力学方程(表 2, R2> 0.94), 故用前4 h的平均脱氮速率代指最大脱氮速率.第1次排泥前, 1号反应器亚硝酸盐去除量为2.10~5.06 mg · L-1, 2号反应器亚硝酸盐去除量为3.43~4.41 mg · L-1.第1次排泥后, 1号反应器亚硝酸盐去除量先降低为1.69 mg · L-1, 然后恢复到3.24 mg · L-1, 最后达到10.37 mg · L-1; 2号反应器亚硝酸盐去除量最大达到5.02 mg · L-1.第2次排泥后, 1号反应器亚硝酸盐去除量先降低为4.56 mg · L-1, 然后恢复到9.58 mg · L-1, 最后达到12.18 mg · L-1; 2号反应器亚硝酸盐去除量最大达到4.24 mg · L-1.结果表明, 排泥之后1号反应器亚硝酸盐去除量较2号反应器明显提高, 证明排泥可以强化反应器脱氮效能.
图 2(Fig. 2)
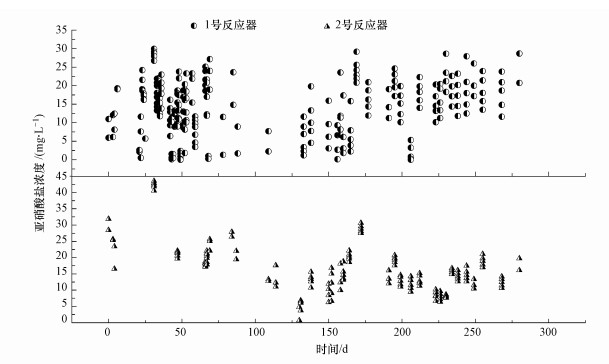 |
| 图 2 亚硝酸盐浓度变化曲线 Fig. 2Concentration variation of nitrite in two reactors |
表 2(Table 2)
| 表 2 反应器排泥前后关键时期脱氮速率 Table 2 Nitrogen removal rate of reactor before and after withdrawing sludge | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
表 2 反应器排泥前后关键时期脱氮速率 Table 2 Nitrogen removal rate of reactor before and after withdrawing sludge
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
两个反应器的脱氮速率如图 3所示.在整个运行过程中, 2号反应器的脱氮速率基本稳定在19 mg · L-1 · d-1.在运行初始阶段, 1号反应器脱氮速率与2号相差不大, 为17.00 mg · L-1 · d-1.第1次排泥后1号反应器脱氮速率逐渐上升至62.21 mg · L-1 · d-1, 为2号反应器的2.23倍.在第2次排泥后, 1号反应器的脱氮速率继续升高, 并在反应末期达到最高值73.10 mg · L-1 · d-1, 是2号反应器的3.41倍.结果表明, 排泥可以提高反应器的脱氮效率.
图 3(Fig. 3)
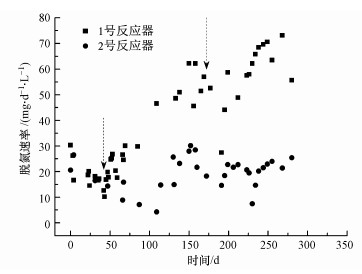 |
| 图 3 脱氮速率变化曲线 (黑色虚线箭头表示排泥) Fig. 3Removal rate of nitrogen in No.1, 2 reactor |
为了进一步探究反应过程, 对两个反应器中的物料进行衡算, 结果如表 3所示.以n-DAMO反应化学计量学为基础, 实测的N2产气速率、甲烷消耗速率均接近于理论值, 表明反应器中发生的是n-DAMO反应, 所以通过排泥可以提高反应器的n-DAMO效能.
表 3(Table 3)
| 表 3 CH4消耗、N2产生速率的理论与实际值对比 Table 3 Theory of production rate of N2, consumption of CH4 compared with the actual value | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
表 3 CH4消耗、N2产生速率的理论与实际值对比 Table 3 Theory of production rate of N2, consumption of CH4 compared with the actual value
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
目前所报道的n-DAMO细菌单独培养脱氮速率及其丰度如表 4所示, 其中, 普遍速率为30~40 mg · L-1 · d-1, 本实验1号反应器在运行到1291 d时脱氮速率达到73.10 mg · L-1 · d-1, 与研究报道的最高速率75.9 mg · L-1 · d-1相差不大, 说明排泥对提高n-DAMO细菌脱氮速率有重要作用.
表 4(Table 4)
| 表 4 n-DAMO过程脱氮速率 Table 4 Nitrogen removal rate of n-DAMO process | ||||||||||||||||||||||||||||||||||||||||||||
表 4 n-DAMO过程脱氮速率 Table 4 Nitrogen removal rate of n-DAMO process
| ||||||||||||||||||||||||||||||||||||||||||||
3.2 排泥对n-DAMO反应器微生物群落结构影响两个反应器中不同阶段的微生物群落结构如图 4所示.通过排泥的操作, 1号反应器内微生物群落结构明显优化, n-DAMO细菌隶属的NC10门所占比例均大幅提高, 从38.3%上升到67.7%.变形菌门、绿弯菌门、绿菌门、芽单胞菌门相对丰度全部降低, 表明排泥可以扩大n-DAMO细菌的生长优势.而2号反应器中NC10相对丰度基本维持不变.其他研究表明, 在经过富集且运行良好的n-DAMO反应器中, n-DAMO细菌丰度一般在50%~80%.本研究初始阶段n-DAMO细菌丰度只有38.3%, 较其他研究偏低, 但经过排泥后n-DAMO细菌丰度提高到67.7%, 与其他研究相当.
图 4(Fig. 4)
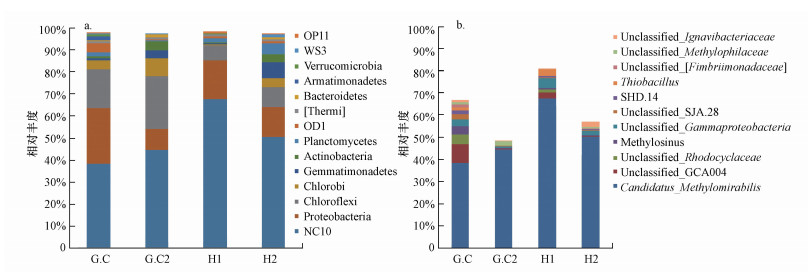 |
| 图 4 门水平(a)和属水平(b)微生物群落结构差异比较 (G.C和H1分别为1号反应器实验132 d、214 d测序结果; G.C2和H2分别为2号反应器实验132 d、214 d测序结果) Fig. 4Comparison of microbial community structure differences at phylum level(a) and genus level(b) |
从属水平上看, 隶属于NC10门的M.oxyfera菌是两个反应器的共同优势微生物.除此之外, 随着排泥的进行大部分在1号反应器中占优的微生物种属, 相对丰度均产生了明显的下降(图 4b).其中, Unclassified_GCA004相对丰度从8.64%下降到2.39%, Unclassified_Rhodocyclaceae相对丰度从4.38%下降到1.43%, Unclassified_Fimbriimonadaceae相对丰度从1.38%下降到0.06%, Methylosinu相对丰度从3.70%下降到0.75%.研究表明, Unclassified_GCA004在微氧条件下有甲烷氧化耦合反硝化的功能(Liu et al., 2013), 通常在红树林沉积物(Wu et al., 2016b)和淡水湖泊沉积物(Wan et al., 2017)中检测到, 在这两种生境下NO3--N和NO2--N浓度分别为0~0.329 mg · L-1、0.063~0.16 mg · L-1(Fernandes et al., 2010); 而在本实验中NO2--N浓度约为30~40 mg · L-1, 且不存在硝酸盐, 适宜n-DAMO细菌的生长(15~50 mg · L-1), 不适宜Unclassified_GCA004的生长, 所以通过排泥可以强化n-DAMO细菌的优势.Yoshida等(2012)在0.3 mg · g-1的稻田土中发现Unclassified_Rhodocyclaceae具有反硝化的能力, 基质利用速率约为0.09 mmol · g-1 · h-1, 而n-DAMO细菌的基质利用速率为0.28 mmol · g-1 · h-1(Allegue et al., 2018), 较Unclassified_Rhodocyclaceae更快, 所以反应器的环境更适宜n-DAMO细菌, 且排泥后n-DAMO细菌基质利用速率更高, 更容易成为优势物种.Methylosinus是一种以甲烷为碳源的甲烷氧化细菌, 研究报道, Methylosinus的甲烷消耗速率为0.95 mg · L-1 · h-1(Ordaz et al., 2014), 而M.oxyfera菌甲烷消耗速率为1.36 mg · L-1 · h-1(Allegue et al., 2018), 说明n-DAMO细菌利用甲烷的能力更强, 排泥使所有微生物同比减少的情况下, n-DAMO细菌凭借高的基质利用速率占有优势.综上所述, 经过排泥后, M.oxyfera菌以其更适合反应器环境与更高的基质利用速率, 使得非目标微生物相对丰度都有所降低, 而M.oxyfera菌丰度增高, 最终达到强化n-DAMO过程的目的.
图 5(Fig. 5)
 |
| 图 5 1号反应器132 d、214 d微生物群落结构差异性分析 Fig. 5Difference analysis of microbial community structure of sludge in No.1 reactor on 132 d and 214 d |
3.3 排泥对功能微生物数量影响反应器中n-DAMO细菌数量如图 6所示.经过排泥, 1号反应器内功能微生物数量大幅提升.qPCR结果表明, 1号反应器132 d n-DAMO细菌的量为1.404×108 copies · g-1, 到214 d增长为4.854×108 copies · g-1, 提高了2.46倍; 2号反应器132 d n-DAMO细菌的量为2.490×108 copies · g-1, 到214 d增长为3.291×108 copies · g-1, 仅增长了32%.在He等(2015b)的研究中, n-DAMO细菌量为(5.0×108±0.4×108) copies · g-1, 与本研究基本一致; 而在Zhou等(2015)的研究中, n-DAMO细菌量为(0.85×108~1.0×108) copies · g-1, 本研究是其1~5倍.实验结果表明, 排泥确实可以直接提高n-DAMO细菌的数量.
图 6(Fig. 6)
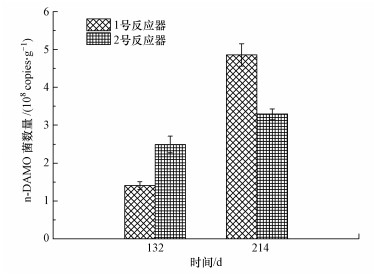 |
| 图 6 反应器中n-DAMO菌绝对数量比较 Fig. 6Comparison of absolute abundance of n-DAMO bacrteria in the reactors |
本文以NO2-为电子受体成功富集n-DAMO功能微生物, 通过排泥对n-DAMO过程进行强化.132 d时1号反应器中n-DAMO细菌数量仅为2号反应器的56.39%, 但其脱氮速率是2号反应器的2.23倍, 表明1号反应器中n-DAMO细菌的活性是2号反应器的3.95倍, 214 d时1号反应器中n-DAMO微生物数量是2号反应器的1.47倍, 脱氮速率达到了2号反应器的3.41倍, 活性提高了2.31倍.1号反应器n-DAMO细菌数量和活性都高于2号反应器, 可能是由于排泥使得n-DAMO细菌丰度提高.1号反应器中n-DAMO细菌丰度所占比例是2号反应器的1.77倍, 达到了67.7%, 与目前已报道的最高丰度相当, 较高的丰度有利于n-DAMO细菌发挥优势作用.
4 结论(Conclusions)1) 排泥能够提高SBR中n-DAMO过程的脱氮速率, 脱氮速率从初期的17.00 mg · L-1 · d-1最高达到73.10 mg · L-1 · d-1, 可满足工程应用需求, 具备开发潜力.
2) 排泥可以选择性富集n-DAMO菌M.oxyfera, 强化后M.oxyfera相对丰度由38.3%上升到67.7%.n-DAMO菌数量由1.404×108 copies · g-1增长到4.854×108 copies · g-1.
3) 排泥后非目标微生物Unclassified_GCA004、Unclassified_Rhodocyclaceae、Unclassified_Fimbriimonadaceae和Methylosinus相对丰度均明显降低, M.oxyfera具有更高的基质利用速度与生长速度可能是其内在原因.
参考文献
| Allegue T, Arias A, Fernandez-Gonzalez N, et al. 2018. Enrichment of nitrite-dependent anaerobic methane oxidizing bacteria in a membrane bioreactor[J]. Chemical Engineering Journal, 347: 721–730.DOI:10.1016/j.cej.2018.04.134 |
| 柴风光, 卢培利, 李微薇, 等. 2018. 利用硝酸盐和亚硝酸盐同步富集厌氧甲烷氧化微生物的比较实验[J]. 微生物学通报, 2018, 45(4): 762–770. |
| Chen J, Zhou Z C, Gu J D. 2014. Occurrence and diversity of nitrite-dependent anaerobic methane oxidation bacteria in the sediments of the South China Sea revealed by amplification of both 16S rRNA and pmoA genes[J]. Applied Microbiology and Biotechnology, 98(12): 5685–5696.DOI:10.1007/s00253-014-5733-4 |
| Ettwig K F, Shima S, Pas-Schoonen K T V D, et al. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea[J]. Environmental Microbiology, 10(11): 3164–3173.DOI:10.1111/emi.2008.10.issue-11 |
| Ettwig K F, Van A T, Kt P S, et al. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum[J]. Applied and Environmental Microbiology, 75(11): 3656–3662.DOI:10.1128/AEM.00067-09 |
| Fernandes S O, Lokabharathi P A, Bonin P C, et al. 2010. Denitrification:an important pathway for nitrous oxide production in tropical mangrove sediments (Goa, India)[J]. Journal of Environmental Quality, 39(4): 1507–1516.DOI:10.2134/jeq2009.0477 |
| Fu L, Ding J, Lu Y Z, et al. 2017. Nitrogen source effects on the denitrifying anaerobic methane oxidation culture and anaerobic ammonium oxidation bacteria enrichment process[J]. Applied Microbiology and Biotechnology, 101(9): 3895–3906.DOI:10.1007/s00253-017-8163-2 |
| Han P, Gu J D. 2013. A newly designed degenerate PCR primer based on pmoA, gene for detection of nitrite-dependent anaerobic methane-oxidizing bacteria from different ecological niches[J]. Applied Microbiology and Biotechnology, 97(23): 10155–10162.DOI:10.1007/s00253-013-5260-8 |
| He Z, Geng S, Shen L, et al. 2015a. The short-and long-term effects of environmental conditions on anaerobic methane oxidation coupled to nitrite reduction[J]. Water Research, 68(47): 554–562. |
| He Z, Cai C, Shen L, et al. 2015b. Effect of inoculum sources on the enrichment of nitrite-dependent anaerobic methane-oxidizing bacteria[J]. Applied Microbiology and Biotechnology, 99(2): 939–946. |
| He Z, Geng S, Wang L, et al. 2016. Improvement of mineral nutrient concentrations and pH control for the nitrite-dependent anaerobic methane oxidation process[J]. Separation and Purification Technology, 162(3): 148–153. |
| Hu B L, Shen L D, Huang Q, et al. 2014a. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands[J]. Proceedings of the National Academy of Sciences of the United States of America, 111(12): 4495–4500.DOI:10.1073/pnas.1318393111 |
| Hu B, He Z, Geng S, et al. 2014b. Cultivation of nitrite-dependent anaerobic methane-oxidizing bacteria:impact of reactor configuration[J]. Applied Microbiology and Biotechnology, 98(18): 7983–7991.DOI:10.1007/s00253-014-5835-z |
| Hu S, Zeng R J, Burow L C, et al. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms[J]. Environmental Microbiology Reports, 1(5): 377–384.DOI:10.1111/j.1758-2229.2009.00083.x |
| Kampman C, Hendrickx T L G, Luesken F A, et al. 2012. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment[J]. Journal of Hazardous Materials, 227: 164–171. |
| Kampman C, Temmink H, Hendrickx T L G, et al. 2014. Enrichment of denitrifying methanotrophic bacteria from municipal wastewater sludge in a membrane bioreactor at 20℃[J]. Journal of Hazardous Materials, 274: 428–435.DOI:10.1016/j.jhazmat.2014.04.031 |
| Li W, Lu P, Chai F, et al. 2018. Long-term nitrate removal through methane-dependent denitrification microorganisms in sequencing batch reactors fed with only nitrate and methane[J]. Amb Express, 8(1): 108.DOI:10.1186/s13568-018-0637-9 |
| Lieberman R L, Rosenzweig A C. 2004. Biological methane oxidation:Regulation, biochemistry, and active site structure of particulate methane monooxygenase[J]. Critical Reviews in Biochemistry and Molecular Biology, 39(3): 147–164.DOI:10.1080/10409230490475507 |
| Liu J, Sun F, Wang L, et al. 2013. Molecular characterization of a microbial consortium involved in methane oxidation coupled to denitrification under micro-aerobic conditions[J]. Microbial Biotechnology, 7(1): 64–76. |
| Lu P, Liu T, Ni B, et al. 2019. Growth kinetics of Candidatus 'Methanoperedens nitroreducens' enriched in a laboratory reactor[J]. Science of the Total Environment, 695: 442–450. |
| Lu Y Z, Fu L, Ding J, et al. 2016. Cr(Ⅵ) reduction coupled with anaerobic oxidation of methane in a laboratory reactor[J]. Water Research, 102: 445–452.DOI:10.1016/j.watres.2016.06.065 |
| Luesken F A, Jaime Sánchez, Alen T A, et al. 2011. Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes[J]. Applied and Environmental Microbiology, 77(19): 6802–6807.DOI:10.1128/AEM.05539-11 |
| Ordaz A, López J C, Figueroa-González I, et al. 2014. Assessment of methane biodegradation kinetics in two-phase partitioning bioreactors by pulse respirometry[J]. Water Research, 67: 46–54.DOI:10.1016/j.watres.2014.08.054 |
| Raghoebarsing A A, Pol A, Pas-Schoonen K T V D, et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification[J]. Nature, 440(7086): 918–921.DOI:10.1038/nature04617 |
| 沈李东, 胡宝兰, 郑平. 2011. 甲烷厌氧氧化微生物的研究进展[J]. 土壤学报, 2011, 48(3): 619–628. |
| Solomon S, Qin M, Manning Z, et al. 2007. Climate Change 2007:The Physical Science Basis[J]. Cambridge University Press: 1–21. |
| Wan Y, Ruan X, Zhang Y, et al. 2017. Illumina sequencing-based analysis of sediment bacteria community in different trophic status freshwater lakes[J]. Microbiology Open, 6: e450.DOI:10.1002/mbo3.450 |
| Wang R, Han X, Lu P, et al. 2016. Co-occurrence and diversity of nitrite-dependent anaerobic methane oxidizing and anaerobic ammonium oxidizing bacteria in the water-level fluctuation zone of the three gorges reservoir, China[J]. Fresenius Environmental Bulletin, 25(12): 5456–5466. |
| Wu Q, Chang J, Yan X, et al. 2016a. Electrical stimulation enhanced denitrification of nitrite-dependent anaerobic methane-oxidizing bacteria[J]. Biochemical Engineering Journal, 106: 125–128.DOI:10.1016/j.bej.2015.11.014 |
| Wu P, Xiong X, Xu Z, et al. 2016b. Bacterial communities in the rhizospheres of three mangrove tree species from Beilun Estuary, China[J]. Plos One, 11(10): e0164082.DOI:10.1371/journal.pone.0164082 |
| Yoshida M, Ishii S, Fujii D, et al. 2012. Identification of active denitrifiers in rice paddy soil by DNA-and RNA-based analyses[J]. Microbes & Environments, 27(4): 456–461. |
| Yue X, Yu G, Liu Z, et al. 2018. Start-up of the completely autotrophic nitrogen removal over nitrite process with a submerged aerated biological filter and the effect of inorganic carbon on nitrogen removal and microbial activity[J]. Bioresource Technology, 254: 347–352.DOI:10.1016/j.biortech.2018.01.107 |
| 赵荣, 朱雷, 吴箐, 等. 2017. 亚硝酸盐型甲烷厌氧氧化过程影响因素研究[J]. 环境科学学报, 2017, 37(1): 178–184. |
| Zhou L, Wang Y, Long X E, et al. 2014. High abundance and diversity of nitrite-dependent anaerobic methane-oxidizing bacteria in a paddy field profile[J]. FEMS Microbiology Letters, 360(1): 33–41.DOI:10.1111/fml.2014.360.issue-1 |
| Zhu B, Van G D, Fritz C, et al. 2012. Anaerobic oxidization of methane in a minerotrophic peatland:enrichment of nitrite-dependent methane-oxidizing bacteria[J]. Applied and Environmental Microbiology, 78(24): 8657–8665.DOI:10.1128/AEM.02102-12 |
