

1. 江南大学 生物工程学院 工业生物技术教育部重点实验室,江苏 无锡 214122;
2. 江南大学 食品安全与营养协同创新中心,江苏 无锡 214122
收稿日期:2018-09-19;接收日期:2018-11-07
基金项目:国家重点研发计划(Nos. 2017YFC1600403, 2017YFC1600405),江苏省重点研发计划(No. BE2016689)资助
摘要:氨基甲酸乙酯(Ethyl carbamate,EC)具有致癌性,广泛存在于酒精饮料中。我国的黄酒因EC含量高而带来的食品安全问题越来越受到人们的关注。微生物酶法消除黄酒中的EC具有直接、高效的特性而被深入研究。文中从黄酒中EC的形成机制、酸性脲酶研究现状、氨基甲酸乙酯水解酶研究现状等方面概述了微生物酶法消除黄酒中EC的研究进展及存在的问题。并针对这些问题,提出了寻找新型氨基甲酸乙酯水解酶、Fe3+依赖型双功能酸性脲酶食品级表达与定向进化及双酶并用将尿素和EC一起消除的策略。
关键词:食品安全氨基甲酸乙酯微生物酶法酸性脲酶氨基甲酸乙酯水解酶黄酒
Advances in microbial enzymatic elimination of ethyl carbamate in Chinese rice wine
Qingtao Liu1,2, Zhen Kang1,2, Guocheng Du1,2


1. The Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu, China;
2. Synergetic Innovation Center of Food Safety and Nutrition, Jiangnan University, Wuxi 214122, Jiangsu, China
Received: September 19, 2018; Accepted: November 7, 2018
Supported by: the National Key Research and Development Program of China (Nos. 2017YFC1600403, 2017YFC1600405), Key Research and Development Program of Jiangsu Province (No. BE2016689)
Corresponding author: Guocheng Du. Tel: +86-510-85918307; Fax: +86-510-85918309; E-mail: gcdu@jiangnan.edu.cn.
Abstract: Ethyl carbamate (EC), a carcinogenic and teratogenic chemical that is widely distributed in various alcoholic beverages, has attracted much attention. Microbial enzymatic degradation of EC in rice wine is always efficient and attractive. In this review, we summarize the research progress and problems of microbial enzymatic elimination of EC in rice wine from three aspects: the mechanisms of EC formation in rice wine, the research progress of acid urease, and the research progress of urethanase. Then, we propose the corresponding strategies to solve the problems: screening new urethanase with satisfied enzyme properties, food-grade expression and directed evolution of the bifunctional Fe3+-dependent acid urease and acid urease used in combination with urethanase to eliminate both urea and EC in rice wine.
Keywords: food safetyethyl carbamateenzymatic eliminationacid ureaseurethanaserice wine
氨基甲酸乙酯(Ethyl carbamate或Urethane,简称EC),是一种具有遗传毒性及较强致癌性的物质[1],天然存在于多种发酵食品(如酱油、食醋、泡菜)和酒精饮料(如黄酒、白酒、葡萄酒等、日本清酒、白兰地)中[2-4]。2007年国际癌症研究机构IARC (the International Agency for Research on Cancer)将EC归类为2A类致癌物质[5]。世界各国和国际卫生组织对酒中的EC浓度都有严格的限量标准[2]。酒精饮料中因EC存在而隐含的食品安全问题越来越受到人们的关注。在酒精饮料中,黄酒的EC含量最高[6],而黄酒基本上都是我国所生产,这不仅限制了我国黄酒的出口,同时也引起人们对其长期饮用所存在的安全方面的担忧。消除黄酒中的EC有多种策略,包括工艺优化、物理吸附、代谢工程策略改造酒用酵母、生物酶法消除等。工艺优化法,一方面通过对原材料的精炼及对发酵过程的控制(如温度、pH控制及添加外源抑制剂等),来减少因原材料带入或菌种代谢及酶促反应所产生的EC的前体物[7-10],另一方面通过对发酵产品的后处理过程进行优化(如缩短灭菌的加热时间、加快灭菌后的降温速度等),来减少EC的形成[11-13]。该过程繁琐,改变生产工艺及采用物理吸附还可能影响酒的风味[3]。代谢工程策略主要是针对酿酒酵母的改造,一方面减少细胞内尿素的生成[14-16],另一方面强化细胞对胞外尿素的吸收及利用[17-19]。该方法仅是通过减少形成EC的前体物质来阻碍EC的形成,对已经形成的EC无法直接消除[20]。而采用微生物酶法可直接降解尿素及EC,其直接、高效的特性使得其在解决酒精饮料因EC而带来的食品安全问题中具有很大的优势[21]。文中从黄酒中EC形成机制、酸性脲酶研究现状、氨基甲酸乙酯水解酶研究现状等方面综述了微生物酶法消除黄酒中EC研究进展及存在的问题,并对未来黄酒中EC酶法消除提出针对性策略。
1 黄酒中氨基甲酸乙酯形成机制EC是由尿素、氨甲酰磷酸、瓜氨酸、焦碳酸二乙酯和氰化物等前体物质与乙醇自发反应而形成[21]。尿素、氨甲酰磷酸、瓜氨酸主要由原材料带入及微生物(酿酒酵母、乳酸菌等)代谢产生,氰化物则主要是在发酵过程中经酶促反应形成,而焦碳酸二乙酯则是作为防腐剂而人为添加[21]。这些前体物质中尿素是黄酒中形成EC的最主要前体物质[3],其次是瓜氨酸[22]。在黄酒的煎酒过程中,尿素、瓜氨酸等前体物质在高温条件下与乙醇快速反应而导致酒中EC大量积累。
黄酒中尿素一方面是由原材料带入,另一方面主要是由酵母细胞在精氨酸代谢过程中产生[23]。精氨酸在精氨酸酶(Arginine hydrolase)作用下水解为鸟氨酸和尿素[24]。一般情况下,尿素再经脲基酰胺酶作用而被分解为氨和CO2。但由于尿素为酵母菌的非偏好氮源,当培养基中存在偏好氮源(如谷氨酰胺及天冬酰胺)时,由于NCR (Nitrogen catabolite repression)效应导致尿素无法被及时分解,进而被尿素转运蛋白转运出细胞,从而大幅增加黄酒中的尿素含量。黄酒中的瓜氨酸主要来自于原材料引入[24],另外瓜氨酸也是由乳酸菌将精氨酸经ADI途径(Arginine deminase pathway)代谢而产生的中间产物[25]。目前为止,对黄酒中EC的消除研究主要集中在消除形成EC的最主要的前体物尿素及直接消除EC上。
2 当前用于消除黄酒中氨基甲酸乙酯的两种酶应用于黄酒中EC消除的酶有两种,其一是酸性脲酶,通过消除黄酒中的尿素来减少EC的形成;另外一种就是氨基甲酸乙酯水解酶,它可以直接降解EC生成氨、乙醇和二氧化碳(图 1)。
 |
| 图 1 酶法消除黄酒中EC Fig. 1 Strategies for enzymatic elimination EC in rice wine. |
| 图选项 |
2.1 酸性脲酶2.1.1 酸性脲酶概述脲酶(Urease, EC 3.5.1.5)是世界上第一个结晶酶[26],可水解尿素生成氨、乙醇及二氧化碳(H2N-CO-NH2 + H2O → H2N-COOH + NH3及H2N-COOH + H2O→H2CO3+ NH3)[27]。脲酶广泛存在于植物、细菌、真菌中,已有很多年的研究历史[28-30]。来自于刀豆的脲酶是第一个被发现的含有Ni2+的金属酶,随后越来越多的脲酶均被证实其活性中心需要Ni2+的参与[31-32]。脲酶由结构亚基(UreA,B,C)及辅助亚基(UreE,F,G,D/H)组成(图 2)。其中来自幽门螺杆菌Helicobacter pylori的脲酶结构基因ureA为其他来源的脲酶ureA与ureB的融合体,其辅助基因ureH的功能与其他来源的脲酶辅助基因ureD的功能一致[33]。脲酶的结构亚基UreC是催化亚基,所有的亚基对于脲酶的活性表达均起到至关重要的作用,缺一不可[30]。
 |
| 图 2 来自于不同微生物的脲酶基因簇 Fig. 2 Structures of urease gene clusters in different microorganisms. |
| 图选项 |
根据脲酶最适作用pH的不同,脲酶分为酸性脲酶、中性脲酶及碱性脲酶[30, 34]。植物来源的脲酶均为中性脲酶,酸性脲酶均发现于微生物中。酸性脲酶因其能在酸性条件下发挥作用,从而能够应用于酒精饮料中尿素的降解而引起了人们的关注。1979年,酸性脲酶在发酵乳杆菌Lactobacillus fermentum中首次被发现[35]。随后许多具有酸性脲酶活力的胃肠道微生物被分离及鉴定[36-37],许多来自于乳酸菌类如嗜热链球菌Streptococcus thermophilus[38-39]、唾液链球菌Streptococcus salivarius[40]、轻型链球菌Streptococcus mitior[41]、罗伊氏乳杆菌Lactobacillus reuteri[42]、L. fermentum[43]等的酸性脲酶也得到了深入的研究。
2.1.2 酸性脲酶应用现状目前来说,来自L. fermentum及运动节杆菌Arthrobacter mobilis的酸性脲酶均已商业化。来自于罗伊氏乳杆菌L. reuteri CICC6124的酸性脲酶,在乳酸乳球菌Lactococcus lactis NZ9000中实现了食品级表达,在3-L发酵罐中通过分阶段控制pH及发酵过程中流加葡萄糖等策略获得了脲酶的产量为11 560 U/L,这是目前所报道的食品级脲酶的最高产量[44]。虽然上述酸性脲酶均能高效降解酒中的尿素,但其均需Ni2+为其配体,而Ni2+的存在引起人们对其应用时安全方面的担忧(表 1)[3-4]。同时,食品级的酸性脲酶产量低(表 1),这也限制了脲酶的工业化应用。近年来,刘庆涛等[45]发现并鉴定了来自于地衣芽孢杆菌Bacillus paralicheniformis 9945A的脲酶为Fe3+依赖型酸性脲酶,其以Fe3+为配体时,在pH 4.5条件下,其Km、Vmax分别为28.2 mmol/L、73.5 μmol/(mg·min)。将此脲酶添加于黄酒中(6 U/mL),在37 ℃条件下反应50 h后可消除92%的尿素(表 1)。此Fe3+依赖型脲酶的发现解除了Ni2+依赖型脲酶应用时安全方面的担忧,在未来工业化应用方面具有重大的潜在价值。
表 1 不同来源的酸性脲酶性质比较Table 1 Comparison of the representative acid urease in different microorganisms
| Source | Ion-dependent | Expression host | Urease activity (U/L) | Urea elimination efficiency | Reference |
| L. fermentum | Nickel | L. fermentum | NP | Addition of urease in sake and reaction at 15 ℃ for 2 d, all the urea were eliminated. | [43] |
| A. mobilis | Nickel | A. mobilis | NP | Addition of 0.09 U/mL urease in sake and reaction at 15 ℃ for 13 d, all the urea were eliminated. | [36] |
| Enterobacter sp. R-SYB082 | Nickel | Enterobacter sp. R-SYB082 | 2 504 | Addition of 0.08 U/mL urease in rice wine and reaction at 37 ℃ for 2 d, 85% urea were eliminated. | [46] |
| L. reuteri CICC6124 | Nickel | L. lactis NZ9000 | 11 560 | Addition of 0.5 U/mL urease in rice wine and reaction at 20 ℃ for 2 h, all the urea were eliminated. | [44, 47] |
| B. paralicheniformis ATCC 9945a | Iron (Fe3+) | E. coli | NP | Addition of 6 U/mL urease in rice wine and reaction at 37 ℃ for 50 h, 92% urea were eliminated. | [45] |
| NP: not report. | |||||
表选项
2.1.3 双功能酸性脲酶的发现杨广明等发现来自于普罗威登斯菌Providencia rettgeri JN-B815的酸性脲酶对EC具有水解作用,研究者克隆出了该酸性脲酶的基因并实现了在大肠杆菌中活性表达[48]。2015年,杨宇清等[44]发现来自于L. reuteri CICC6124的酸性脲酶对EC具有水解作用,通过对黄酒中EC的降解分析发现,该酶虽然耐乙醇、耐酸,但对黄酒中的EC几乎无直接降解作用。可能的原因是脲酶对EC的底物亲和力(Km)差,导致其对黄酒中低浓度的EC无法降解。刘庆涛等[45]发现来自于B. paralicheniformis 9945A的Fe3+依赖型脲酶对EC也具有水解作用。在pH 4.5条件下,该酶对EC的Km、Vmax分别为958 mmol/L、15.2 μmol/(mg·min),如此低的EC亲和力及催化效率导致其不能有效降解黄酒中的EC。同时刘庆涛等也发现普罗威登斯菌Providencia sp. LBBE的脲酶对EC也具有水解作用。研究者通过染色体步移技术[49],从该菌基因组上获得了脲酶基因簇的核苷酸序列,并实现了其在大肠杆菌中活性表达[45]。具有EC降解能力的双功能酸性脲酶的发现,为黄酒中EC降解提供了新的思路。
2.1.4 酸性脲酶应用于黄酒中EC消除时存在的问题应用酸性脲酶虽然能够高效消除黄酒中的尿素,从而减少EC的形成。但由于尿素并非形成EC的唯一前体物质,且其对已经形成的EC无法降解,因而此种方法仅能减少黄酒中由尿素形成的EC;双功能酸性脲酶虽然对EC有水解作用,但其对EC的底物亲和力差、催化效率低等问题致使其不能直接降解黄酒中低浓度的EC;另外食品级宿主所生产的脲酶产量低,也限制了其应用。
2.1.5 酸性脲酶未来研究方向酸性脲酶未来的研究方向,一方面集中在对双功能酸性脲酶的底物特异性改造上。来自于B. paralicheniformis的酸性脲酶,因其为Fe3+依赖型脲酶,且耐酸、耐乙醇并能够降解黄酒中的尿素而具有改造价值。脲酶的结构亚基UreC为催化亚基,包含了脲酶的催化活性中心。利用同源建模技术对C亚基进行3D结构模拟,利用Discovery studio等软件将底物EC对接到UreC的活性口袋。通过分子动力学模拟校正对接结果。通过分子对接结果,对脲酶C亚基进行半理性设计改造,从而提高其对EC的底物亲和力及催化效率(图 3)。
 |
| 图 3 脲酶UreC亚基同源建模及分子对接结构图(A:UreC 3-D结构图;B:脲酶催化活性中心;C:脲酶底物结合口袋) Fig. 3 Homology modeling and molecular docking structure of UreC. (A) 3-D structure of UreC. (B) Catalytic sites of UreC. (C) Substrate binding pocket of UreC. |
| 图选项 |
另一方面,酸性脲酶未来的研究方向集中在对酸性脲酶的食品级高效制备上。提高脲酶在食品级宿主中的表达量,有以下几点建议:其一,优化脲酶基因簇各个基因间的核糖体结合位点(RBS),确保所有基因均表达。脲酶基因簇由结构基因及辅助基因构成(图 2),各个基因间存在交互重叠的现象,这样就会导致异源表达整个基因簇时,翻译的过程有可能会意外终止。而任何一个基因的不表达均能导致脲酶失去活力[30]。其二,强化催化亚基UreC的表达,从而平衡脲酶结构亚基UreA、B、C之间的表达量。文献报道,在同一个操纵子中表达多个基因时,距离启动子越远的基因表达量越低[50]。而大部分脲酶均以(UreABC)3的组成结构发挥催化作用,UreC表达量的减弱会造成UreA、B的浪费,因此,有必要强化UreC的表达。其三,与Ni2+或Fe3+转运蛋白共表达。文献报道,Ni2+依赖型的脲酶与Ni2+转运蛋白共表达可显著提高脲酶的比酶活力[51-52]。因此与Fe3+转运蛋白共表达有可能会提高Fe3+依赖型脲酶异源表达时的酶活力。
2.2 氨基甲酸乙酯水解酶2.2.1 氨基甲酸乙酯水解酶概述氨基甲酸乙酯水解酶(英文名称为urethane hydrolase或urethane amidohydrolase,urethanase),酶学委员会编号EC 3.5.1.75。日本****最早从事EC水解酶的研究。1990年Kobashi等[53]在柠檬酸杆菌Citrobacter sp.中发现EC水解酶,并证实其功能为降解EC生成氨、二氧化碳及乙醇,由此便命名其为“urethanase”。因其所发现的EC水解酶只能水解酰胺及氨基甲酸酯,而对有机酸酯没有任何水解作用,他们推测EC水解酶属于酰胺酶家族,并沿用至今。
目前,尚没有EC水解酶水解EC机制的报道。从EC分子结构来看,其存在酰胺键(R-CO-NH-R)和酯键(R-COO-R),理论上来说,酰胺酶及酯酶家族中的部分酶类均能对EC有水解作用(图 4)。但是,目前为止,已报道的具有EC降解能力的酶均属于酰胺酶家族酶类,尚未有酯酶水解EC的报道。
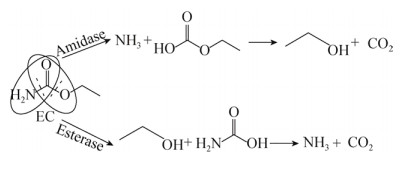 |
| 图 4 EC水解机制 Fig. 4 The mechanism of EC hydrolysis. |
| 图选项 |
2.2.2 氨基甲酸乙酯水解酶的研究现状EC水解酶来源广泛,酶学性质差异较大(表 2)。具有代表性的EC水解酶有两个:其一是2006年Akutsu-Shigeno等[54]从马红球菌(Rhodococcus equistrain TB-60)中所发现并克隆的EC水解酶(GenBank: DD320008.1)。该酶在大肠杆菌中获得了的活性表达。酶学性质分析发现,该酶虽然在酸性条件下可以降解EC,但对EC的底物亲和力差(Km=6.59 mmol/L),催化效率低(kcat/Km < 0.01 L/(mol·s)且在乙醇存在条件下不稳定,导致其无法应用于酒中EC的降解[54]。其二是李京京等[55]从小鼠胃中筛选出一株具有EC降解能力的赖氨酸芽孢杆菌Lysinibacillus fusiformis SC02。通过对此菌株所产的EC水解酶进行蛋白纯化及N端测序,并基于N端测序信息,设计简并引物,扩增得到了EC水解酶的基因序列(GenBank Accession No. KU353448.1),最终在大肠杆菌及枯草芽孢杆菌中实现了该酶的活性表达。但该酶乙醇耐受性差,在5%的乙醇中即失去活力,同时酸耐受性差,在低于pH 6.0的环境中不稳定等特性限制了其应用。随后,刘晓慧等[55]将此EC水解酶在大肠杆菌中进行了定向进化的改造。通过计算机辅助设计的方法,获得了温度稳定性提高的突变体,但该突变体的乙醇耐受性及耐酸性并没有太大的变化,依然远不能达到其在黄酒中应用的需求。其他来源的EC水解酶基因序列均未解析。
表 2 不同来源的EC水解酶性质比较Table 2 Comparison of urethanase properties from different sources
| Source | Optical pH | Relative activity (%) | Km (mmol/L) | Reference |
| Citrobacter sp. | 6.0–8.0 | 8.9a | 1.600 | [53] |
| Bacillus licheniformis sp.1013 | 4.5 | 54a | 42.000 | [56] |
| Bacillus licheniformis IFO 12107 | 4.5 | 64a | 0.170 | [57] |
| Micrococcus species | 4.5 | 82a | 0.078 | [58] |
| Rhodococcus equi TB-60 | 5.5 | NP | 6.590 | [54] |
| Rhodotorula mucilaginosa LBMAE-8 | NP | 40b | NP | [59] |
| Penicillium variabile JN-A525 | 6.0 | 65b | 27.200 | [60] |
| Klebsiella pneumoniae | 7.5 | 10a | 74.000 | [61] |
| Lysinibacillus fusiformis SCO2 | 7.0 | 10b | 37.200 | [55] |
| The enzyme activity that measured in the absence of ethanol was considered as 100%. a: enzyme activity was measured in the presence of 20% (V/V) ethanol; b: enzyme activity was measured in the presence of 15% (V/V) ethanol; NP: not report. | ||||
表选项
2.2.3 氨基甲酸乙酯水解酶应用于黄酒中EC消除存在的问题EC水解酶应用于黄酒中EC消除时存在以下问题:其一,EC水解酶的乙醇耐受性及酸耐受性差,难以在含有高浓度乙醇及酸性条件下保持稳定(表 2)。其二,EC水解酶对EC的底物亲和力及催化效率低,难以降解黄酒中痕量的EC (100–750 μg/L)[6],同时由于EC与尿素分子结构相近,有些能够水解EC的酰胺酶对尿素也有水解作用,导致尿素与EC之间存在底物竞争。由于黄酒中尿素含量(10–50 mg/L)是EC含量的100–1 000倍左右,可能会造成尿素对EC竞争性抑制而使得EC水解酶不能发挥作用。其三,EC水解酶氨基酸序列尚未得到有效解析,目前仅2个EC水解酶编码序列得以鉴定。这些缺点导致目前所发现的EC水解酶难以实际应用于黄酒中EC的降解。
2.2.4 氨基甲酸乙酯水解酶未来研究方向目前所鉴定出基因序列的两个EC水解酶不耐乙醇及酸,致使其无法在黄酒中使用,因此筛选新型EC水解酶至关重要。由于能够水解EC的酰胺酶对尿素可能也有水解作用,可能导致黄酒中存在的尿素对EC竞争性抑制,因此未来EC水解酶筛选方向有两种:其一,筛选对尿素无水解作用的耐乙醇耐酸的酰胺酶;其二,筛选对EC具有水解作用的耐乙醇耐酸的酯酶。另外筛选方法上,也应该从传统的筛选EC水解酶产生菌转移到利用生物信息学手段,从酶库中直接筛选具有EC水解能力的酶。笔者已从150多个酯酶中筛选到了一个耐乙醇且能降解EC的酯酶,后续将对其作出应用评价。
3 展望随着人们生活水平的提高,食品安全问题越来越受到人们的关注。如何消除酒中具有致癌性的EC,已经成为一个世界性问题。目前为止,微生物酶法(酸性脲酶及EC水解酶)在应用于黄酒中EC及其前体物尿素的消除中,具有直接、高效的优势。然而目前所发现的EC水解酶在酸性或乙醇存在条件下不稳定以及酸性脲酶对EC底物亲和力低、产量低、Ni2+作为辅助因子而带来的食品安全性问题,使得其尚无法在黄酒中应用。针对这些问题,我们对黄酒中EC的酶法降解提出了以下3个策略:1)筛选新型EC水解酶,即筛选对尿素无水解作用的耐乙醇耐酸的酰胺酶或筛选对EC具有水解作用的耐乙醇耐酸的酯酶。2) Fe3+依赖型双功能酸性脲酶的底物特异性改造及食品级高效表达。一方面通过蛋白质工程技术对双功能酸性脲酶进行定向进化,提高其对EC的亲和力及催化效率;另一方面,通过食品级表达系统高效制备改造后的双功能酸性脲酶。3)酸性脲酶与EC水解酶并用彻底消除黄酒中的EC。黄酒煎酒前,使用酸性脲酶消除尿素,减少在煎酒过程中EC的形成,同时也减少了成品黄酒在储存过程中EC的再次形成;煎酒后,使用EC水解酶(或改造后的双功能酸性脲酶)对已经形成的EC直接消除(目前所发现的EC水解酶暂时不能实现黄酒中EC的直接消除),从而解决黄酒因EC而存在的食品安全问题(图 1)。
参考文献
| [1] | Forkert PG. Mechanisms of lung tumorigenesis by ethyl carbamate and vinyl carbamate.Drug Metab Rev, 2010, 42(2): 355–378.DOI: 10.3109/03602531003611915 |
| [2] | Weber JV, Sharypov VI. Ethyl carbamate in foods and beverages: a review.Environ Chem Lett, 2009, 7(3): 233–247.DOI: 10.1007/s10311-008-0168-8 |
| [3] | Zhao XR, Du GC, Zou HJ, et al. Progress in preventing the accumulation of ethyl carbamate in alcoholic beverages.Trends Food Sci Technol, 2013, 32(2): 97–107.DOI: 10.1016/j.tifs.2013.05.009 |
| [4] | Gowd V, Su HM, Karlovsky P, et al. Ethyl carbamate: an emerging food and environmental toxicant.Food Chem, 2018, 248: 312–321.DOI: 10.1016/j.foodchem.2017.12.072 |
| [5] | Lachenmeier DW. Consequences of IARC re-evaluation of alcoholic beverage consumption and ethyl carbamate on food control.Deut Lebensm Rundsch, 2007, 103: 307–311. |
| [6] | Chen DW, Ren YP, Zhong QD, et al. Ethyl carbamate in alcoholic beverages from China: levels, dietary intake, and risk assessment.Food Control, 2017, 72: 283–288.DOI: 10.1016/j.foodcont.2015.10.047 |
| [7] | Yoshizawa K, Takahashi K, Sato K. Changes of urea content in rice and sake moromi during sake making process.Nippon Jozo Kyokaishi, 1988, 83: 136–141. |
| [8] | Liu SQ, Pilone GJ. A REVIEW: arginine metabolism in wine lactic acid bacteria and its practical significance.J Appl Microbiol, 1998, 84(3): 315–327.DOI: 10.1046/j.1365-2672.1998.00350.x |
| [9] | Hasnip S, Caputi A, Crews C, et al. Effects of storage time and temperature on the concentration of ethyl carbamate and its precursors in wine.Food Addit Contam, 2004, 21(12): 1155–1161.DOI: 10.1080/02652030400019851 |
| [10] | Fang RS. The metabolism mechanism and inhibition method of ethyl caramate formation during traditional Chinese rice wine fermentation[D]. HangZhou: Zhejiang University, 2017 (in Chinese). 方若思.传统黄酒发酵中氨基甲酸乙酯产生的代谢规律及抑制方法研究[D].杭州: 浙江大学, 2017.http://journals.im.ac.cn/html/cjbcn/2019/4/%20//cdmd.cnki.com.cn/Article/CDMD-10335-1017071368.htm |
| [11] | Wu HM, Chen L, Pan GS, et al. Study on the changing concentration of ethyl carbamate in yellow rice wine during production and storage by gas chromatography/mass spectrometry.Eur Food Res Technol, 2012, 235(5): 779–782.DOI: 10.1007/s00217-012-1807-7 |
| [12] | Park SR, Ha SD, Yoon JH, et al. Exposure to ethyl carbamate in alcohol-drinking and nondrinking adults and its reduction by simple charcoal filtration.Food Control, 2009, 20(10): 946–952.DOI: 10.1016/j.foodcont.2009.02.006 |
| [13] | Liu J, Zhao GA, Xu Y. Directly removal of ethylcarbamate in Chinese rice wine.J Food Sci Biotechnol, 2012, 31(2): 171–176.(in Chinese). 刘俊, 赵光鳌, 徐岩. 黄酒中氨基甲酸乙酯直接减除技术的研究.食品与生物技术学报, 2012, 31(2): 171-176.DOI:10.3969/j.issn.1673-1689.2012.02.009 |
| [14] | Suizu T, Iimura Y, Gomi K, et al. Construction of urea non-producing yeast Saccharomyces cerevisiae by disruption of the CAR1 gene.Agric Biol Chem, 1990, 54(2): 537–539. |
| [15] | Kitamoto K, Oda K, Gomi K, et al. Genetic engineering of a sake yeast producing no urea by successive disruption of arginase gene.Appl Environ Microbiol, 1991, 57(1): 301–306. |
| [16] | Wu DH, Li XM, Shen C, et al. Decreased ethyl carbamate generation during Chinese rice wine fermentation by disruption of CAR1 in an industrial yeast strain.Int J Food Microbiol, 2014, 180: 19–23.DOI: 10.1016/j.ijfoodmicro.2014.04.007 |
| [17] | Wu D, Li X, Lu J, et al. Constitutive expression of the DUR1, 2 gene in an industrial yeast strain to minimize ethyl carbamate production during Chinese rice wine fermentation.FEMS Microbiol Lett, 2016, 363(1): fnv214.DOI: 10.1093/femsle/fnv214 |
| [18] | Coulon J, Husnik JI, Inglis DL, et al. Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine.Am J Enol Viticult, 2006, 57(2): 113–124. |
| [19] | Dahabieh MS, Husnik JI, van Vuuren HJJ. Functional enhancement of Sake yeast strains to minimize the production of ethyl carbamate in Sake wine.J Appl Microbiol, 2010, 109(3): 963–973.DOI: 10.1111/jam.2010.109.issue-3 |
| [20] | Zhang P, Hu X. Metabolic engineering of arginine permeases to reduce the formation of urea in Saccharomyces cerevisiae.World J Microbiol Biotechnol, 2018, 34: 47.DOI: 10.1007/s11274-018-2430-y |
| [21] | Mohapatra BR. An Insight into the prevalence and enzymatic abatement of urethane in fermented beverages//Patra JK, Das G, Shin HS, eds. Microbial Biotechnology. Singapore: Springer Singapore, 2018: 153-170. |
| [22] | Wu DH, Li XM, Sun JY, et al. Effect of citrulline metabolism in Saccharomyces cerevisiae on the formation of ethyl carbamate during Chinese rice wine fermentation.J Inst Brew, 2018, 124(1): 77–84.DOI: 10.1002/jib.v124.1 |
| [23] | An D, Ough CS. Urea excretion and uptake by wine yeasts as affected by various factors.Am J Enol Viticult, 1993, 44(1): 35–40. |
| [24] | Schehl B, Senn T, Lachenmeier DW, et al. Contribution of the fermenting yeast strain to ethyl carbamate generation in stone fruit spirits.Appl Microbiol Biotechnol, 2007, 74(4): 843–850.DOI: 10.1007/s00253-006-0736-4 |
| [25] | Azevedo Z, Couto JA, Hogg T. Citrulline as the main precursor of ethyl carbamate in model fortified wines inoculated with Lactobacillus hilgardii: a marker of the levels in a spoiled fortified wine.Lett Appl Microbiol, 2002, 34(1): 32–36.DOI: 10.1046/j.1472-765x.2002.01045.x |
| [26] | Sumner JB. The isolation and crystallization of the enzyme urease. Preliminary paper.J Biol Chem, 1926, 69(2): 435–441. |
| [27] | Krajewska B. Ureases Ⅰ. functional, catalytic and kinetic properties: a review.J Mol Catal B: Enzym, 2009, 59(1/3): 9–21. |
| [28] | Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization.Microbiol Rev, 1989, 53(1): 85–108. |
| [29] | Balasubramanian A, Ponnuraj K. Crystal structure of the first plant urease from Jack Bean: 83 years of journey from its first crystal to molecular structure.J Mol Biol, 2010, 400(3): 274–283. |
| [30] | Carter EL, Flugga N, Boer JL, et al. Interplay of metal ions and urease.Metallomics, 2009, 1(3): 207–221.DOI: 10.1039/b903311d |
| [31] | Dixon NE, Gazzola C, Blakeley RL, et al. Jack bean urease (EC 3.5.1.5). Metalloenzyme. Simple biological role for nickel.J Am Chem Soc, 1975, 97(14): 4131–4133.DOI: 10.1021/ja00847a045 |
| [32] | Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter.J Biol Chem, 2013, 288(19): 13178–13185.DOI: 10.1074/jbc.R112.446526 |
| [33] | Fong YH, Wong HC, Yuen MH, et al. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease.PLoS Biol, 2013, 11(10): e1001678.DOI: 10.1371/journal.pbio.1001678 |
| [34] | McDonald JA, Vorhaben JE, Campbell JW. Invertebrate urease: purification and properties of the enzyme from a land snail, Otala lactea.Comp Biochem Physiol Part B: Comp Biochem, 1980, 66(2): 223–231.DOI: 10.1016/0305-0491(80)90056-5 |
| [35] | Suzuki K, Benno Y, Mitsuoka T, et al. Urease-producing species of intestinal anaerobes and their activities.Appl Environ Microbiol, 1979, 37(3): 379–382. |
| [36] | Miyagawa K, Sumida M, Nakao M, et al. Purification, characterization, and application of an acid urease from Arthrobacter mobilis.J Biotechnol, 1999, 68(2/3): 227–236. |
| [37] | Yang LQ, Wang SH, Tian YP. Purification, properties, and application of a novel acid urease from Enterobacter sp.Appl Biochem Biotechnol, 2010, 160(2): 303–313.DOI: 10.1007/s12010-008-8159-6 |
| [38] | Mora D, Maguin E, Masiero M, et al. Characterization of urease genes cluster of Streptococcus thermophilus.J Appl Microbiol, 2004, 96(1): 209–219.DOI: 10.1046/j.1365-2672.2003.02148.x |
| [39] | Zotta T, Ricciardi A, Rossano R, et al. Urease production by Streptococcus thermophilus.Food Microbiol, 2008, 25(1): 113–119.DOI: 10.1016/j.fm.2007.07.001 |
| [40] | Chen YY, Clancy KA, Burne RA. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque Streptococcus.Infect Immun, 1996, 64(2): 585–592. |
| [41] | Yamazaki E, Kurasawa T, Kakimoto S, et al. Characteristics of acid urease from Streptococcus mitior.Agric Biol Chem, 1990, 54(9): 2433–2435. |
| [42] | Kakimoto S, Sumino Y, Akiyama SI, et al. Purification and characterization of acid urease from Lactobacillus reuteri.Agric Biol Chem, 1989, 53(4): 1119–1125. |
| [43] | Kakimoto S, Sumino Y, Kawahara K, et al. Purification and characterization of acid urease from Lactobacillus fermentum.Appl Microbiol Biotechnol, 1990, 32(5): 538–543. |
| [44] | Yang YQ, Kang Z, Zhou JL, et al. High-level expression and characterization of recombinant acid urease for enzymatic degradation of urea in rice wine.Appl Microbiol Biotechnol, 2015, 99(1): 301–308.DOI: 10.1007/s00253-014-5916-z |
| [45] | Liu QT, Chen YQ, Yuan ML, et al. A Bacillus paralicheniformis iron-containing urease reduces urea concentrations in rice wine.Appl Environ Microbiol, 2017, 83(17): e01258–17. |
| [46] | Liu J, Xu Y, Nie Y, et al. Optimization production of acid urease by Enterobacter sp. in an approach to reduce urea in Chinese rice wine.Bioprocess Biosyst Eng, 2012, 35(4): 651–657.DOI: 10.1007/s00449-011-0643-7 |
| [47] | Zhou JL, Kang Z, Liu QT, et al. Degradation of urea and ethyl carbamate in Chinese Rice wine by recombinant acid urease.Chin J Biotech, 2016, 32(1): 74–83.(in Chinese). 周建立, 康振, 刘庆涛, 等. 重组酸性脲酶对黄酒中尿素和氨基甲酸乙酯的降解应用.生物工程学报, 2016, 32(1): 74-83. |
| [48] | Liu XF, Zhang Q, Zhou ND, et al. Expression of an acid urease with urethanase activity in E. coli and analysis of urease gene.Mol Biotechnol, 2017, 59(2/3): 84–97. |
| [49] | Wang SM, He J, Cui ZL, et al. Self-formed adaptor PCR: a simple and efficient method for chromosome walking.Appl Environ Microbiol, 2007, 73(15): 5048–5051.DOI: 10.1128/AEM.02973-06 |
| [50] | Juminaga D, Baidoo EEK, Redding-Johanson AM, et al. Modular engineering of L-tyrosine production in Escherichia coli.Appl Environ Microbiol, 2012, 78(1): 89–98. |
| [51] | Bauerfeind P, Garner RM, Mobley LT. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity.Infect Immun, 1996, 64(7): 2877–2880. |
| [52] | Chen YYM, Burne RA. Identification and characterization of the nickel uptake system for urease biogenesis in Streptococcus salivarius 57.I.J Bacteriol, 2003, 185(23): 6773–6779.DOI: 10.1128/JB.185.23.6773-6779.2003 |
| [53] | Kobashi K, Takebe S, Sakai T. Urethane-hydrolyzing enzyme from Citrobacter sp.Chem Pharm Bull, 1990, 38(5): 1326–1328.DOI: 10.1248/cpb.38.1326 |
| [54] | Akutsu-Shigeno Y, Adachi Y, Yamada C, et al. Isolation of a bacterium that degrades urethane compounds and characterization of its urethane hydrolase.Appl Microbiol Biotechnol, 2006, 70(4): 422–429.DOI: 10.1007/s00253-005-0071-1 |
| [55] | Liu XH, Fang F, Xia XL, et al. Stability enhancement of urethanase from Lysinibacillus fusiformis by site-directed mutagenesis.Chin J Biotech, 2016, 32(9): 1233–1242.(in Chinese). 刘晓慧, 方芳, 夏小乐, 等. 定点突变改造提高纺锤形赖氨酸芽孢杆菌氨基甲酸乙酯水解酶稳定性.生物工程学报, 2016, 32(9): 1233-1242. |
| [56] | Zhao CJ, Imamura L, Kobashi K. Urethanase of Bacillus licheniformis sp. isolated from mouse gastrointestine.Chem Pharm Bull, 1991, 39(12): 3303–3306.DOI: 10.1248/cpb.39.3303 |
| [57] | Zhao C, Kobashi K. Purification and characterization of iron-containing urethanase from Bacillus licheniformis.Biol Pharm Bull, 1994, 17(6): 773–778.DOI: 10.1248/bpb.17.773 |
| [58] | Mohapatra B, Bapuji M. Characterization of urethanase from Micrococcus species associated with the marine sponge (Spirasfrella species).Lett Appl Microbiol, 1997, 25(6): 393–396.DOI: 10.1111/lam.1997.25.issue-6 |
| [59] | Wu Q, Zhao Y, Wang D, et al. Immobilized Rhodotorula mucilaginosa: a novel urethanase-producing strain for degrading ethyl carbamate.Appl Biochem Biotechnol, 2013, 171(8): 2220–2232.DOI: 10.1007/s12010-013-0493-7 |
| [60] | Zhou ND, Gu X, Tian Y. Isolation and characterization of urethanase from Penicillium variabile and its application to reduce ethyl carbamate contamination in chinese rice wine.Appl Biochem Biotechnol, 2013, 170(3): 718–728.DOI: 10.1007/s12010-013-0178-2 |
| [61] | Bu PP, Chen J, Du GC. Purification and characterization of a halophilic urethanase from Klebsiella pneumoniae.Chin J Biotech, 2014, 30(12): 404–411.(in Chinese). 卜攀攀, 陈坚, 堵国成, 等. 耐盐氨基甲酸乙酯水解酶的分离纯化及酶学性质.生物工程学报, 2014, 30(12): 404-411. |
