影响因子:10.812
刊物名称:Molecular Plant
出版年份:4 May 2020
https://doi.org/10.1016/j.molp.2020.03.011.
Abstract
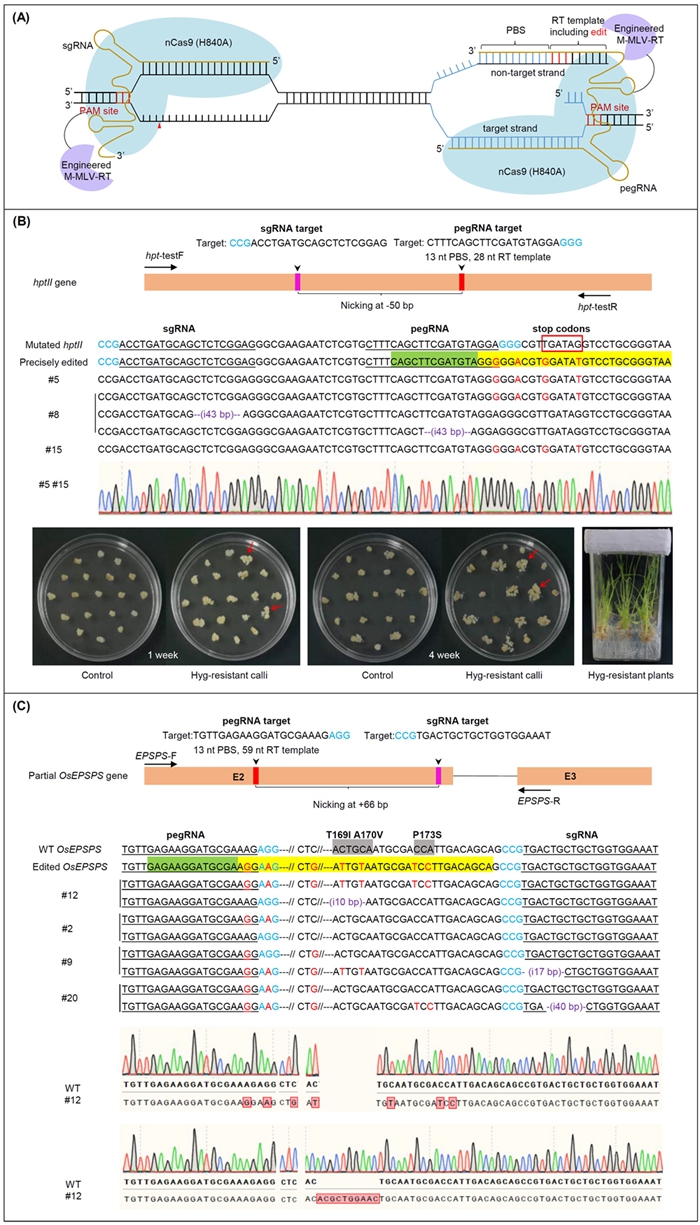
Harnessing genetic diversity and the introduction of elite alleles from wild relatives or landraces into commercial cultivars has been a major goal in crop breeding programs. Precise modification of the plant genome through clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) (CRISPR/Cas)-mediated homology-directed repair (HDR) offers agreat promise to introduce elite alleles from wild relatives or landraces into commercialized cultivars in the short term. Various strategies have been attempted for precise targeted gene/allele replacement or gene insertion and tagging through CRISPR/Cas systems in plants (Baltes etal., 2014,Gil-Humanes etal., 2016,Sun etal., 2016,Wang etal., 2017,Li etal., 2019,Miki etal., 2018,Hua etal., 2019;Wolter and Puchta, 2019,Dong etal., 2020). However, due to the intrinsic lower frequency of HDR in plant cells, insufficient availability of a donor repair template, and the non-homologous end-joining (NHEJ) is the predominant DNA repair pathway in plants (Baltes etal., 2014,Li etal., 2019), it remains challenging, especially in crop plants. Thus, it is essential to further exploit more efficient precision genome-editing technology in order to accelerate crop improvement.